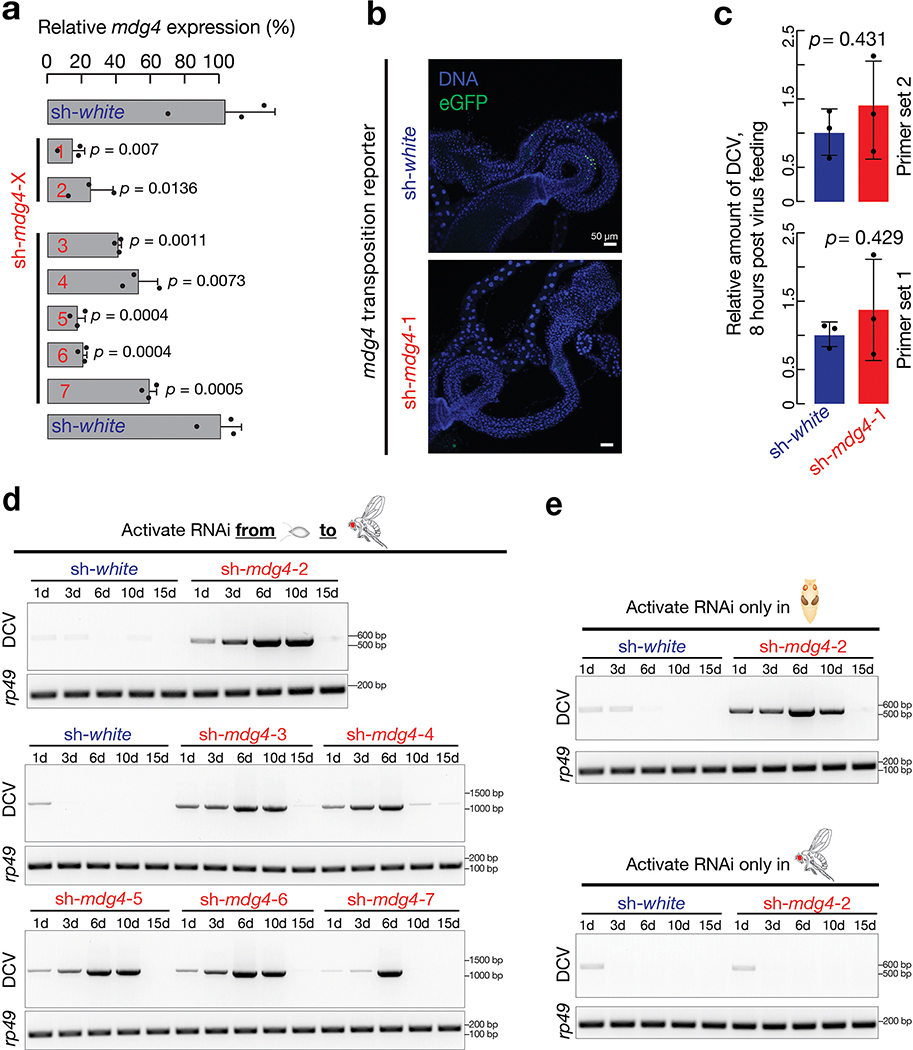
Extended Data Fig. 5 |. Multiple RNAi constructs were designed to silence mdg4.
a, RT-qPCR to quantify the expression of mdg4 upon suppression by using one of the 7 RNAi constructs based on the two-tailed t-test. Each sh-RNA construct was driven by ac-Gal4. Flies were raised at 25°C and newly eclosed flies were used to extract RNA. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates (apply to all RT-qPCR data from this manuscript). b, Visualizing mdg4 transposition events (GFP positive cells) in hindgut from either white or mdg4 suppressed 2–4-day-old adults. Flies carrying sh-mdg4-5 construct were used for Fig. 6. sh-mdg4-1 was used in the rest of the experiments. The findings from it were validated by using other constructs (shown in Extended Data Fig. 11). c, RT-qPCR to measure the amount of DCV from fly bodies after feeding animals virus for 8 hours based on the two-tailed t-test. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates. d, By activating mdg4 RNAi from embryonic to adult stage, RT-PCR experiments were performed to monitor the amount of DCV in fly bodies after one-time infection. Together with Extended Data Fig. 5a, these data indicate RNAi efficiency negatively correlates with the robustness of antiviral response. e, By using sh-mdg4-2 to silence mdg4 at the stage specific manner, similar findings were made as Fig. 5. Three independent biological replicates were performed for sh-mdg4-2 in d and e, one time experiment was performed for sh-mdg4-3, 4, 5, 6 and 7 in d.