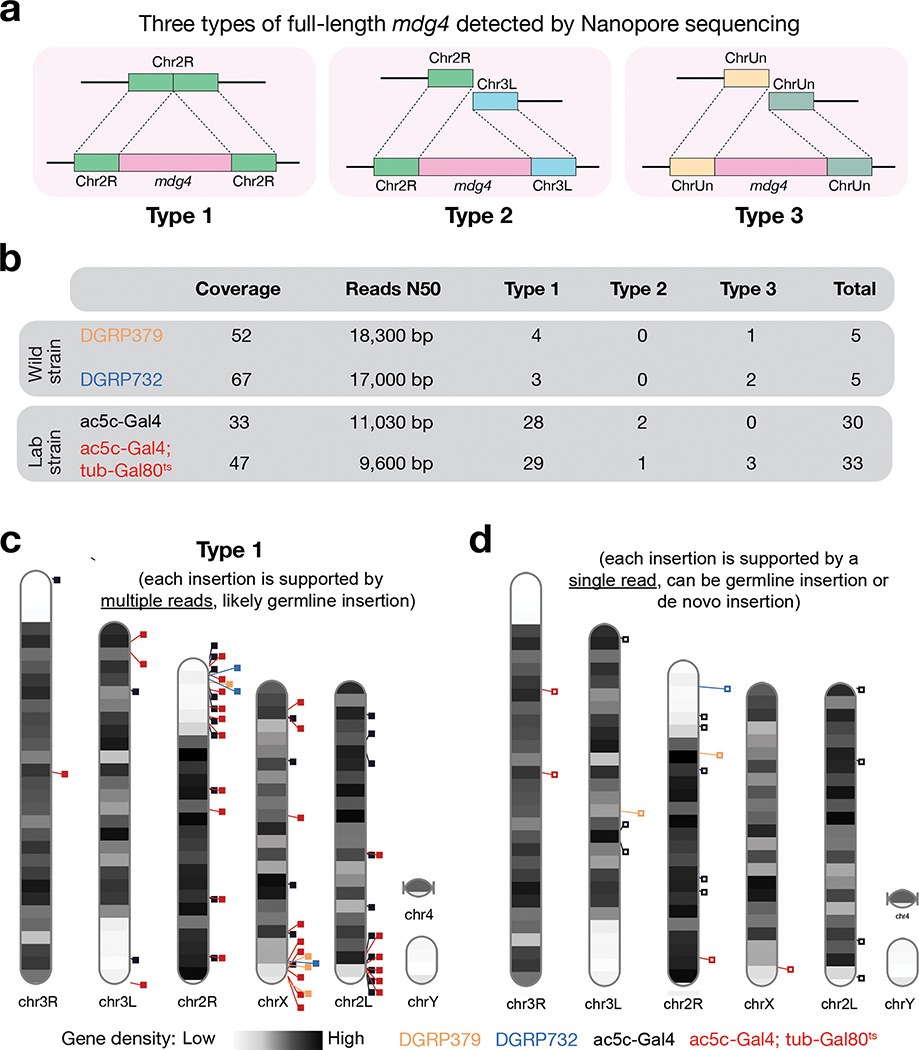
Fig. 3 |. Nanopore sequencing detected full-length copies of mdg4 from both wild and laboratory strains.
a, Cartoon display of 3 types of full-length mdg4 detected. Type 1: Full-length mdg4 flanked by coordinated sequences from the reference genome. Type 2: Full-length mdg4 flanked by discordant sequences from the reference genome, likely reflecting the polymorphisms between the reference genome and the genome of individual strain. Type 3: Full-length mdg4 flanked unassembled genome sequences. b, A table to summarize the sequencing depth, N50 of the sequencing reads, and the number of full-length mdg4 detected from each strain. c, Chromosome ideogram to display the chromosomal positions of type 1 full-length mdg4. Each event reported in panels b and c is supported multiple reads, likely reporting germline mdg4 copies. d, Chromosome ideogram to display the chromosomal positions of full-length mdg4 detected by a single read, likely reporting de novo insertions or germline insertions with low regional coverage.