FIGURE 3.
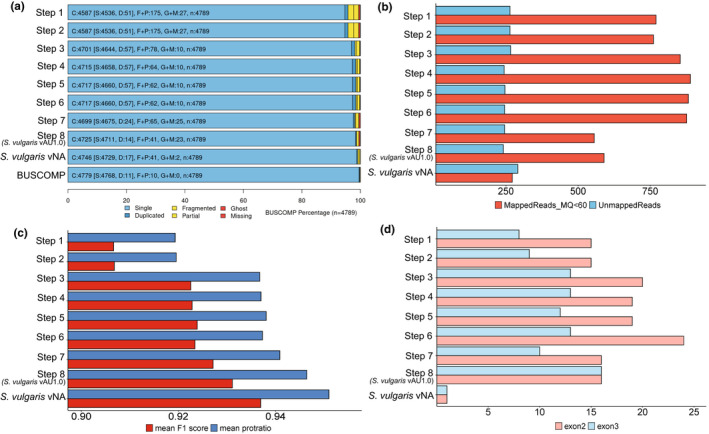
Sturnus vulgaris vAU assembly steps overview. Quality and completeness assessments for eight sequential assembly steps: Step 1 (supernova assembly), step 2 (diploidocus primary assembly), step 3 (sspace‐longreads scaffolding), step 4 (gapfinisher gapfilling), step 5 (L_rna_scaffolder), step 6 (pilon polishing), step 7 (diploidocus clean up), and step 8 (satsuma2 chromosome scaffolding; S. vulgaris vAU1.0), and S. vulgaris vNA. (a) buscomp completeness results for the 4779 busco genes identified as single copy and complete in one or more assembly stages. The final buscomp row compiles the best rating for each gene across all eight steps (complete: 95% + coverage in a single contig/scaffold; duplicated: 95% + coverage in 2+ contigs/scaffolds; fragmented: 95% + combined coverage but not in any single contig/scaffold; partial: 40%–95% combined coverage; ghost: Hits meeting local cutoff but <40% combined coverage; missing: No hits meeting local cutoff). (b) a bar plot of the number of Iso‐Seq reads that for each assembly step failed to map (blue) or fell below a mapQ score of 60 (red). (c) saaga annotation scores of mean protein length ratio (blue) and F1 score (red) (see methods for details). (d) Total count of MHCIIB exon 2 (red) and exon3 (blue) sequences identified in each assembly
