Abstract
Introduction
Brown seaweeds are a sustainable biomass with a potential for various industrial applications. Polyphenols are an important contributor to this potential.
Objective
The aim was total quantification of polyphenols in brown seaweeds from different tidal zones, using a selective 1H quantitative NMR (qNMR) method, comparing the results with the colorimetric Folin–Ciocalteu total phenolic content (TPC) assay.
Method
qNMR was performed with integration of selected peaks in the aromatic region (7–5.5 ppm). Deselection of non‐polyphenolic 1H signals was based on information from 2D (1H‐13C, 1H‐15N) NMR spectra. 13C NMR phlorotannin characterisation facilitated the average number of protons expected to be found per aromatic ring used for the 1H quantification.
Results
Selective qNMR and the TPC assay showed similar results for the three sublittoral growing species from the Laminariaceae; lower amounts for Laminaria hyperborea and Laminaria digitata (qNMR: 0.4%–0.6%; TPC: 0.6%–0.8%, phloroglucinol equivalents (PGE), dry weight (DW)) and higher amounts for Saccharina latissima (qNMR: 1.2%; TPC: 1.5%, PGE, DW). For the eulittoral Fucaceae, Fucus vesiculosus (qNMR: 1.1%; TPC: 4.1%; PGE, DW) and Ascophyllum nodosum (qNMR: 0.9%; TPC: 2.0%; PGE, DW), the TPC results were found to be up to three times higher than the qNMR results. The 13C NMR characterisation showed the highest phlorotannin polymerisation degree for F. vesiculosus .
Conclusion
The TPC assay provided similar polyphenolic amounts to the selective qNMR method for sublittoral species. For eulittoral growing species, the TPC method showed amounts up to three times higher than the qNMR method—most likely illustrating the lack of selectivity in the TPC assay.
Keywords: phlorotannins, polyphenols, qNMR, quantification, seaweeds
Short abstract
An optimised 1H qNMR method was used for quantification of polyphenols in seaweeds by utilising 13C NMR phlorotannin characterisation and selective 1H integration based on 2D NMR data. qNMR results were compared with a colourimetric TPC assay, showing similar results for sublittoral brown seaweeds with a similar phlorotannin polymerisation degree. The eulittoral growing species, having a different phlorotannin polymerisation, showed TPC results up to three times the qNMR results—most likely illustrating the shortcomings of the TPC assay.
1. INTRODUCTION
The world's population is projected to reach 9 billion by 2050, and utilisation of bioresources will be increasingly important for food, feed, and health applications. Seaweeds, or macroalgae, are a sustainable biomass and play an increasing role in aquaculture and marine bioresource development. 1 , 2 , 3 , 4 Macroalgae grow in abundance in their natural habitat and can be both harvested and farmed. Seaweed farming is a sustainable industry with minimal environmental impact, as it does not require fertilisers or irrigation and does not compete for agricultural land. 1 , 5 , 6 , 7 , 8 , 9 Several products, for various applications, can be extracted from macroalgae including alginate, fucoidan, mannitol, cellulose, proteins, carotenoids, and polyphenols. 10 , 11
Polyphenols are bioactive compounds synthesised by macroalgae during plant growth and as a response to external stressors such as UV radiation, wounding, and climate. 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 Bioactivities of polyphenols include antioxidant, antiviral, anticancer, antibacterial, antidiabetic, and neuroprotective activities as well as antiallergic effects. 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 Various polyphenols have been identified in macroalgae, with phlorotannins being the predominant polyphenol group in the class of Phaeophyta (brown algae). 12 , 36 , 37 , 38 , 39 , 40 , 41 , 42
Phlorotannins, which are exclusive to brown algae, are oligomers of phloroglucinol and are separated into different subgroups depending on the linkage of the phloroglucinol units. These linkages can be either phenyl linkages (C‐C), ether linkages (C‐O‐C), or both (Figure 1). 9 , 12 , 18 , 43 , 44 , 45
FIGURE 1.
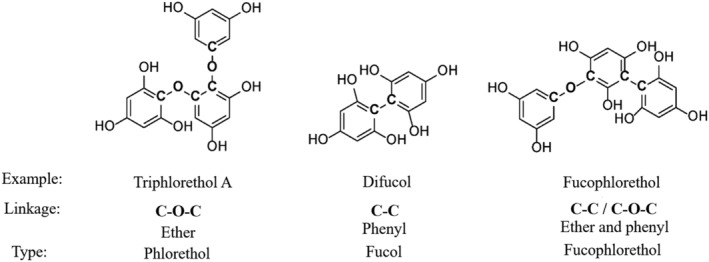
Examples of phlorotannins containing the different phloroglucinol linkage types triphlorethol A (ether linkage, C‐O‐C, phlorethol type), difucol (phenyl linkage, C‐C, fucol type), and fucophlorethol (ether and phenyl linkage, fucophlorethol type)
Extensive analysis and identification of polyphenols in algae is important to uncover and understand algae's potential in industrial applications. However, existing methods for polyphenolic analysis have significant shortcomings, such as lack of exactness at the molecular level. This is due to the low concentrations of polyphenols in the large compound matrix of the algae. 36 , 46 , 47 Furthermore, the diversity of seaweed polyphenols complicates data collection and standardisation of procedures. 9 Being able to quantify the polyphenolic content in crude seaweed materials with higher accuracy is important in order to fully explore seaweeds' potential applications. 48 , 49 , 50 , 51 Table 1 displays selected literature with variation in methods, standards, and polyphenolic amounts found for different species representing three different brown seaweed families; Fucaceae, Sargassaeae, and Laminariaceae.
TABLE 1.
Selected reported quantifications of polyphenols (PP) from brown seaweeds found for species within the Fucaceae, Sargassaceae, and Laminariaceae families, utilising either the TPC assay or qNMR, indicating variation in reference standards used
| Seaweed | Location | Extraction solvent | Quantification method | PP concentration | Publication | |
|---|---|---|---|---|---|---|
| Fucaceae | ||||||
| Fucus vesiculosus | Denmark | Ethanol | TPC | 12.0 mg GAE/g * | Farvin and Jacobsen (2013) 52 | |
| F. vesiculosus | France | Ethanol | qNMR | 15.32% TAE | Parys et al. (2007) 53 | |
| F. vesiculosus | France | Ethanol | TPC | 15.88% PGE | Parys et al. (2007) 53 | |
| F. vesiculosus | Ireland | 60% aqueous methanol | TPC | 2.5 mg GAE/g DW | O'Sullivan et al. (2011) 54 | |
| F. vesiculosus | Canada | 50% aqueous methanol | TPC | 23.21% PGE | Zhang et al (2006) 55 | |
| F. vesiculosus | Iceland | 70% aqueous acetone | TPC | 242 mg PGE/g * | Wang et al. (2009) 56 | |
| Fucus serratus | Ireland | 80% ethanol | TPC | 0.075 mg GAE/g * | Heffernan et al. (2014) 57 | |
| F. serratus | Ireland | 70% aqueous acetone | TPC | 30.68 mg PGE/g | Ford et al. (2020) 45 | |
| F. serratus | Ireland | 70% aqueous acetone | qNMR | 17.00 mg TAE/g | Ford et al. (2020) 45 | |
| Ascophyllum nodosum | Ireland | 80% ethanol | TPC | 0.101 mg PGE/g * | Tierney et al. (2013) 58 | |
| A. nodosum | Spain | Water | TPC | 59.2 mg PGE/g DW | Gisbert et al. (2021) 59 | |
| A. nodosum | Ireland | 70% aqueous acetone | TPC | 36.68 mg PGE/g | Ford et al. (2020) 45 | |
| A. nodosum | Ireland | 70% aqueous acetone | qNMR | 37.35 mg TAE/g | Ford et al. (2020) 45 | |
| A. nodosum | Scotland | Ethanol | TPC | 0.3%–1.0% PGE FW | Parys et al. (2009) 60 | |
| A. nodosum | Scotland | Ethanol | qNMR | 0.6%–2.2% TAE FW | Parys et al. (2009) 60 | |
| A. nodosum | France | Ethanol | TPC | 13.49% PGE | Parys et al. (2007) 53 | |
| A. nodosum | France | Ethanol | qNMR | 25.34% TAE | Parys et al. (2007) 53 | |
| Sargassaceae | ||||||
| Sargassum muticum | France | 75% ethanol | TPC | 10.18% PGE | Anaëlle et al. (2013) 61 | |
| Sargassum fusiforme | China | 30% aqueous ethanol | TPC | 63.61 mg PGE/g | Li et al. (2017) 62 | |
| Cystoseira tamariscifolia | France | 50% aqueous methanol | TPC | 0.63% PGE | Jégou et al. (2015) 63 | |
| C. tamariscifolia | France | 50% aqueous methanol | qNMR | 0.46% PGE | Jégou et al. (2015) 63 | |
| Laminariaceae | ||||||
| Macrocystis pyrifera | Chile | 70% aqueous acetone | TPC | 1.47 mg GAE/g DW | Leyton et al. (2016) 64 | |
| Laminaria hyperborea | Ireland | 60% aqueous methanol | TPC | 1.5 mg GAE/g DW | O'Sullivan et al. (2011) 54 | |
| L. hyperborea | Iceland | 70% aqueous acetone | TPC | 130 mg PGE/g * | Wang et al. (2009) 56 | |
| Laminaria digitata | Iceland | 70% aqueous acetone | TPC | 10 mg PGE/g * | Wang et al. (2009) 56 | |
| L. digitata | Denmark | Ethanol | TPC | 0.324 mg GAE/g * | Farvin and Jacobsen (2013) 52 | |
| L. digitata | Scotland | 80% aqueous methanol | TPC | 5.7% GAE | Vissers et al. (2017) 44 | |
| L. digitata | Scotland | 80% aqueous methanol | qNMR | 4.3% GAE | Vissers et al. (2017) 44 | |
| L. digitata | Ireland | 80% ethanol | TPC | 0.0022 mg GAE/g * | Heffernan et al. (2014) 57 | |
| Saccharina latissima | Canada | 50% aqueous methanol | TPC | 2.17% PGE | Zhang et al. (2006) 55 | |
| Saccharina latissima | Norway | 80% aqueous acetone | TPC | 5–15 mg PGE/g DW | Roleda et al. (2019) 65 | |
Value recalculated to mg (GAE/PGE)/g from original publication.
Abbreviations: DW, dry weight; GAE, gallic acid equivalents; PGE, phloroglucinol equivalents; TAE, trimesic acid equivalents; TPC, total phenolic content.
The Folin–Ciocalteu (FC) total phenolic content (TPC) colorimetric assay was introduced nearly 100 years ago and is still the most used method for polyphenol quantification. 66 However, the method depends on a non‐selective redox reaction and has been evaluated to yield only estimates of polyphenol content. 36 , 67 , 68 , 69 , 70 High performance liquid chromatography (HPLC) with UV‐visible diode array detection (DAD) is also used for polyphenol quantification but rarely for quantification of the total polyphenolic content. This is due to the quantification method being based on molar absorptivity (ε; Beer–Lambert's law, A = εcl). Molar absorptivity (ε) values vary greatly, even within polyphenol classes, making a precise “one standard” total polyphenolic quantification with HPLC‐DAD impossible. 71 Quantitative NMR is a quantification method independent of colorimetric changes, molar absorptivity, and calibration curves. 36 , 44 , 45 , 53 , 72 , 73 , 74 The method quantifies polyphenols based on correlations between signals of polyphenols and an internal or external standard significantly different from the analyte. However, the method should not be used without some knowledge of the polyphenolic nature of the extract and reasonable selection of NMR peaks for quantification. 36
In this study, we continue our examination of total phenolic quantification methods for seaweeds by optimising the quantitative 1H NMR method and compare the results with the FC TPC assay. 36 Three brown seaweeds from the Laminariaceae, Laminaria hyperborea, Laminaria digitata, Saccharina latissima (syn. Laminaria saccharina), and two Fucaceae species, Ascophyllum nodosum and Fucus vesiculosus, were selected for the examination. The selected Laminariaceae species are distributed in the sublittoral zone, while the Fucaceae species have their natural habitat from the middle littoral to lower intertidal zone: the eulittoral zone. Thus, the Fucaceae species are more exposed to greater variation in environmental conditions such as solar radiation (UV) and temperature fluctuation than the Laminariaceae species, possibly reflected in their polyphenol content. 75 This study's main objective was to advance toward optimised quantification tools for analysis of polyphenols in seaweeds to increase the accuracy of these assessments. 13C NMR was used to assess the different linkage profiles of the phlorotannins in the examined brown seaweed species.
2. MATERIALS AND METHODS
2.1. Chemicals
All chemicals used were of analytical grade. The FC reagent, gallic acid, phloroglucinol, methanol (≥ 99.9%), ethanol (absolute), ethyl acetate (≥ 99.5%), DMSO2 (TraceCERT®), and DMSO‐d 6 (0.03% TMS) were acquired from Sigma‐Aldrich (Sigma‐Aldrich, St. Louis, MO, USA). Deionised water was deionised at the University of Bergen (Bergen, Norway).
2.2. Seaweed material
Laminaria hyperborea leaves were acquired from Alginor ASA. Samples were harvested in March 2020 (M20), September 2020 (S20), and August 2021 (A21) along the coast of Haugesund, Norway (Rogaland field 55E; N 59°11′ E 005°06′). Laminaria digitata leaf samples were also acquired from Alginor ASA. The material was collected in August 2019 along the southern Australian coast, Melbourne, Victoria. Fucus vesiculosus samples were collected from Storåkervika, Bergen, Norway (N 60°30.1044′ E 5°15.6726′) in August 2019. Saccharina latissima (syn. L. saccharina) was acquired from Lerøy AS. The material was harvested outside Trollsøy, Vestland (N 60°8.42′ E 5°14.88′) in June 2021. Ascophyllum nodosum was collected in Eidsvåg, Bergen (N 60°26.63′ E 05°17.87′) in September 2017. All samples were rinsed thoroughly with fresh water and air dried. The plant material was stored at −20°C when not used.
2.3. Sample preparation
Crude extracts of each macroalgae were obtained using similar extraction parameters to the ones established by Ummat et al (2020). 76 A total of 10–20 g of dried material was pre‐soaked with water (500 mL) for 30 min in an ultrasound bath (35 kHz). The same material was further extracted with aqueous ethanol (50:50, v/v; 2 x 500 mL) in the ultrasound bath for 30 min. All extractions of the same material were pooled and dried for analysis. When not used, dried crude extracts were stored at −20°C.
2.4. Folin–Ciocalteu TPC assay
Procedures described by Singleton et al. (1999) and Singleton and Rossi (1965) with slight modifications optimised for brown seaweeds were used to determine the TPC using the FC reagent. 77 , 78 Briefly, in the method 0.2 mL sample, blank or standard, 1.59 mL FC reagent, and 4.0 mL 20% (w/v) Na2CO3 were used and made to a total volume of 20 mL with water. The mixture was incubated for 2 h in the dark, and absorbance was measured at 760 nm using a Biochrom Libra S32 UV instrument (Biochrom, Cambridge, United Kingdom). Gallic acid and phloroglucinol calibration curves were used to validate the linearity, sensitivity, precision, and accuracy of the TPC method (Table 2). Three parallels (n = 3) of each sample or standard were analysed to ensure statistically significant results.
TABLE 2.
Calibration curve, limit of detection (LOD) and limit of quantification (LOQ) for gallic acid and phloroglucinol at 760 nm using the optimal total phenolic content (TPC) conditions
| Standard | Calibration curve | r 2 | Range [ug/mL] | LOD [ug/mL] | LOQ [ug/mL] |
|---|---|---|---|---|---|
| Gallic acid | y = 0.00115x – 0.00101 | 0.999 | 1000–30 | 18.610 | 56.391 |
| Phloroglucinol | y = 0.00102x – 0.00177 | 0.998 | 1000–30 | 44.067 | 133.54 |
2.5. NMR analyses
Dried samples were dissolved in 0.6 mL DMSO‐d 6 (0.03% TMS) containing the internal standard DMSO2 (C = 10mM). Quantification using 1H NMR analyses was performed employing a Bruker 600 MHz instrument (Bruker BioSpin, Zürich, Switzerland). All spectra were recorded at 298 K. For accurate quantification, the T1 value of each sample was measured to ensure complete relaxation between scans. The T1 measurements were performed by applying the t1ir pulse sequence with a sweep width of 19.8 ppm, 16 k data points, 8 scans, 4 dummy scans, and 9 different inversion recovery delays between 1 ms and 5 s. To ensure complete relaxation, the d1 value was set to 5 × T1 for all 1H spectra obtained for quantitative NMR (qNMR) analysis. 36 , 79
The one‐dimensional (1D) 1H NMR spectra used for quantifications were recorded using the zg30 pulse sequence with a sweep width of 19.8 ppm, 65 k data points, 128 scans, 2 dummy scans, and the relaxation delay (d1) was 5 × T1 for the selected sample. The spectra were processed using a line broadening of 0.3 Hz.
All quantifications were performed based on Equation 1 with DMSO2 (10mM, No. H = 6, MW = 94.13 g/mol) as the internal standard.
| (1) |
where C = molar concentration [M], I = signal integral, and n = number of protons yielding the signal.
Aromatic signals in the region of 7.0–5.5 ppm were individually integrated and quantified and then added together to obtain the estimated TPC. Standard samples of gallic acid and phloroglucinol were analysed, integrated, and quantified to yield the standard deviation of the qNMR method (Table 3).
TABLE 3.
Standard deviations of the qNMR method meaured with two standards, gallic acid and phloroglucinol, with known concentrations (Cknown) using DMSO2 as the internal reference
| Standard | C known [M] | Chemical shift [ppm] | Integral | Number of protons (n) | C measured [M] | Standard deviation |
|---|---|---|---|---|---|---|
| Gallic acid | 0.0355 | 6.91 | 1.18 | 2 | 0.0353 | 0.000115 |
| Phloroglucinol | 0.0362 | 5.70 | 2.36 | 3 | 0.0471 | 0.00544 |
Two‐dimensional (2D) 1H‐13C and 1H‐15N NMR spectra (heteronuclear multiple‐bond correlation (HMBC) and heteronuclear single‐quantum coherence (HSQC)) were used to eliminate non‐aromatic signals in the polyphenol region (7.0–5.5 ppm).
1H‐13C HMBC spectra were acquired using the hmbcetgpl3nd pulse sequence with non‐uniform sampling (50%), 352 scans, 16 dummy scans, 1H sweep width of 13.02 ppm, 13C sweep width of 220.0 ppm, and a relaxation time of 2.0 seconds.
1H‐13C HSQC spectra used the hsqcedetgpsisp2.3 pulse sequence with 128 scans, 32 dummy scans, 1H sweep width of 13.02 ppm, and 13C sweep width of 200.0 ppm.
Additionally, 1H‐15N HSQC spectra were recorded utilising the hsqcetgp pulse sequence. Number of scans was 32 with 8 dummy scans, 1H sweep width of 15.15 ppm, and 15N sweep width of 200.0 ppm.
1D 13C NMR spectra were used qualitatively to identify linkage differences of phlorotannins in the samples. Spectra were recorded using the udeft pulse sequence with 21 k data points, 236.65 ppm sweep width, 5,120 scans, and 8 dummy scans. The udeft pulse sequence was used, so more sensitive 13C spectra with maximised signal‐to‐noise ratio could be acquired in shorter time. 80 , 81 Signals of 95–160 ppm were used to obtain carbon ratios distinguishing characteristic phlorotannin carbons.
3. RESULTS AND DISCUSSION
3.1. Phlorotannin characterisation with 13C NMR
As phlorotannins are the dominating polyphenolic compounds in brown seaweeds, 13C NMR was used to assess the linkage profiles of the phlorotannins in the five seaweeds examined (Table 4). 13C NMR spectra of L. hyperborea (M20), L. digitata, S. latissima, A. nodosum, and F. vesiculosus were interpreted based on predicted chemical shifts and literature data. 44 , 45 Figure 2 displays the 13C NMR spectra of the examined species (A‐E) and indicates the characteristic signal regions for typical phlorotannin linkages. The two carbons of the phlorotannin ether linkages (C‐O‐C) were observed between 124 and 128 ppm and 156 and 161 ppm in the 13C NMR spectrum, while signals from phenyl linkages (C‐C) were found between 100 and 105 ppm. Signals representing the C‐H bonds in the aromatic phlorotannin were found between 96 and 99 ppm. To make an overall characterisation of the phlorotannin content present for each species, the relative occurrence of ether linkages (C‐O‐C) and phenyl linkages (C‐C) in the 13C NMR spectrum can be compared. The measured intensities of the different linkage signals are calculated relative to the aromatic phlorotannin C‐H carbon: I (C‐H) = 1 (Table 4). 44 , 45
TABLE 4.
Measured intensity (13C NMR) of characteristic phlorotannin linkages in the examined seaweed species presented relative to each species’ aromatic C‐H carbon
| Intensity ratio measured | |||
|---|---|---|---|
| Species | C‐O‐C | C‐C | C‐H a |
| Laminaria hyperborea M20 | 3.08 | 1.00 | 1.00 |
| Laminaria digitata | 0.94 | 0.32 | 1.00 |
| Saccharina latissima | 2.73 | 0.68 | 1.00 |
| Fucus vesiculosus | 20.1 | 1.11 | 1.00 |
| Ascophyllum nodosum | 4.95 | 5.67 | 1.00 |
I(C‐H) = 1.0; C‐O‐C = ether linkage; C‐C = phenyl linkage.
FIGURE 2.
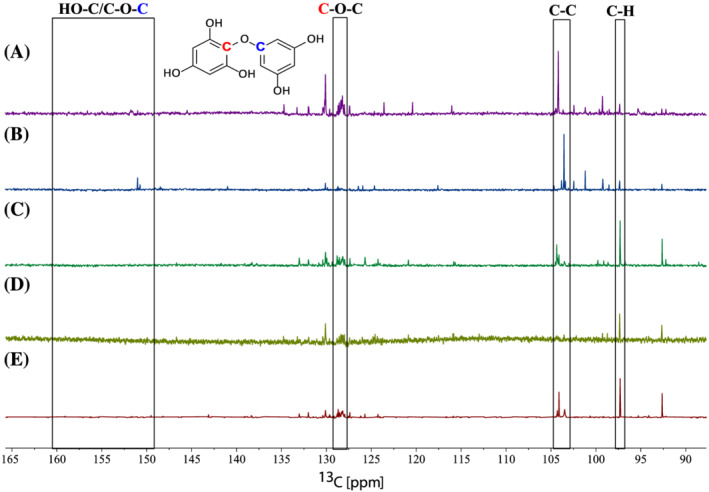
13C NMR spectra of Fucus vesiculosus (A), Ascophyllum nodosum (B), Saccharina latissima (C), Laminaria digitata (D), and Laminaria hyperborea M20 (E) demonstrating the structural linkage differences. The boxes indicate the peaks representing ether linkages (C‐O‐C), phenyl linkages (C‐C), and the C‐H bonds. A hypothetical structure is drawn to indicate the difference in chemical shifts of the two carbons in the ether linkages (C‐O‐C). Figure 1 illustrates the different phlorotannin linkages.
Ford et al. (2020) and Vissers et al. (2017) report on characterisation of phlorotannins in brown seaweeds using 13C NMR data. A. nodosum and Fucus serratus in the study by Ford et al. (2020) were both found to be dominated by phlorethol‐like (ether linkage) phlorotannins. 45 Vissers et al. (2017) report a higher abundance of ether linkages compared with phenyl linkages in L. digitata. 44 In the same study, they also present a molar fucol‐to‐phlorethol ratio of 1:26, which means that for each phenyl linkage there are 26 ether linkages within the phlorotannins in the extract, indicating an abundance of phlorethol‐like phlorotannins. The intensity data from our analysis (Table 4) indicate that phlorotannins with ether linkages are more abundant compared with those with phenyl linkages in four of the five seaweeds in this study (L. hyperborea, L. digitata, S. latissima, and F. vesiculosus). Of these, F. vesiculosus shows, by far, the highest fucol‐to‐phlorethol ratio (1:18). The calculated linkage ratio of these species indicates a larger presence of phlorethol‐type phlorotannins compared with fucol‐type phlorotannins (Figure 1, Table 4). The fifth brown alga, A. nodosum, showed a distinct ratio of an approximate equal occurrence of phenyl linkages compared with ether linkages (1:0.8), in accordance with fucophlorethol‐type phlorotannins. However, an even distribution of the two linkages, resulting in a similar ratio, is also a possibility. Although Ford et al. (2020) did not report a fucol‐to‐phlorethol ratio, calculations based on the data provided in their study yield the fucol‐to‐phlorethol ratio is 1:7 for A. nodosum and 1:2 for F. serratus. The linkage ratios are, to some extent, sample specific because the intensities of each characteristic carbon signal are measured relative to the aromatic C‐H carbon of that selected sample. This will cause results to vary, but the differences could also reflect both seasonal and geographical variation in the seaweeds’ phlorotannin content. However, the dissimilar results might also reflect the natural variation of the polyphenolic content. 82 , 83 , 84 Overall, the presented data indicate that brown algae contain more phlorethol‐like phlorotannins (Table 4), supporting the reports by Visser et al and Ford et al and reinforced by mass spectrometry analyses previously reported. 41 , 85 , 86
Furthermore, the phlorotannin 13C NMR characterisation can facilitate an estimate of the number of protons per aromatic phlorotannin ring in the seaweed samples. This estimate is made to improve the accuracy of the total polyphenolic content quantification using 1H qNMR. 44 , 53 phlorethols (Figure 1) generally consist of one terminal aromatic ring containing three aromatic hydrogens, yielding signals in the characteristic polyphenol region of the 1H spectrum, whereas the remaining phloroglucinol units only have two aromatic hydrogens. In fucols, the terminal units contain two aromatic hydrogens and the internal unit(s) only one. Additionally, when a phloroglucinol unit is connected to ≥ 3 subunits (polymerisation degree ≥ 4), the average number of aromatic hydrogens decreases. 44 The species' polymerisation degree is investigated to some extent, as the ratios of ethyl (C‐O‐C) and phenyl (C‐C) linkages are reported relative to the aromatic carbon (C‐H). Table 4 indicates that the Laminariaceae have the lowest polymerisation degree, with L. digitata having the lowest. Both A. nodosum and F. vesiculosus show high degree of polymerisation with either I(C‐O‐C) or I(C‐C) well above 3. Additionally, other studies have found indications that seaweeds belonging to the Fucaceace species contain phlorotannins consisting of 2–16 phloroglucinol units. 41 , 85 , 87 , 88 Montero et al. (2016) found that for the brown algae Sargassum muticum, the degree of polymerisation of the phlorotannins ranged from 2 to 10 for samples collected in Norway. 89 Taking these studies and the indications of the relative intensity ratios measured into account, it can be assumed that phlorotannins with polymerisation degrees ≥ 4 make up a large part of the phlorotannin matrix of the alga in this study. Considering the 13C ratios and the knowledge of the expected structures, the average number of protons per aromatic ring was set to be 2H (nsample = 2, Equation 1). The number of hydrogens present per aromatic ring estimates the number of protons available per polyphenol in the sample, and thus this educated assumption of the number of protons in the samples was used in the qNMR calculations (Table 5).
TABLE 5.
Total polyphenol content obtained for Laminaria hyperborea, Laminaria digitata, Saccharina latissima, Ascophyllum nodosum, and Fucus vesiculosus extracts using the selective qNMR method. Results are expressed as both gallic acid equivalents (GAE) and phloroglucinol equivalents (PGE) per dry weight (DW).
| Sample | Family | Zone | C [mg GAE/g DW] | C [mg PGE/g DW] |
|---|---|---|---|---|
| L. hyperborea M20 | Laminariaceae | Sublittoral | 8.32 ± 0.00 | 6.17 ± 0.01 |
| L. hyperborea S20 | Laminariaceae | Sublittoral | 5.51 ± 0.00 | 4.08 ± 0.01 |
| L. hyperborea A21 | Laminariaceae | Sublittoral | 6.57 ± 0.00 | 4.87 ± 0.01 |
| L. digitata | Laminariaceae | Sublittoral | 6.86 ± 0.00 | 5.09 ± 0.01 |
| S. latissima | Laminariaceae | Sublittoral | 16.8 ± 0.0 | 12.4 ± 0.0 |
| A. nodosum | Fucaceae | Eulittoral | 11.57 ± 0.0 | 8.57 ± 0.0 |
| F. vesiculosus | Fucaceae | Eulittoral | 14.8 ± 0.0 | 11.0 ± 0.0 |
3.2. Total quantification of polyphenols
1H qNMR can be performed by integrating the ‐OH spectral region (14–8 ppm), as proposed by Nerantzaki et al. (2011). 74 More conventional methods, however, integrate the aromatic region (8–6 ppm). 36 , 44 , 53 , 60 , 63 , 72 , 73 Due to possible H‐D exchange with aromatic–OH groups, leading to loss of intensity and broad peaks, the aromatic 1H‐region was selected for polyphenolic quantification. Based on knowledge of chemical 1H‐shifts of polyphenolic aromatic signals, a narrower region (7.0–5.5 ppm) was selected in order not to integrate signals from the same aromatic system twice. 44 , 45 , 53 Two‐dimensional NMR spectra (HMBC and HSQC) of the seaweed extracts were analysed to explore the nature of the proton signals in the defined region followed by a selective peak‐picking process prior to integration. For example, all samples revealed a similar peak around 6 ppm in the proton spectra (Figure 4). 1H‐15N HSQC spectra indicated that this 1H peak was coupled to a nitrogen δ 5.97/81.8 (1H/15N), meaning this signal is unlikely to originate from the polyphenolic biosynthesis (Figure 3). Therefore, this peak was not quantified. Similar peak picking was performed based on recorded 1H‐13C HMBC spectra for each alga. Figure 4 displays the quantified region of the 1H NMR spectrum for the five algae with eliminated signals indicated. Signals were individually integrated and quantified (Equation 1), then summed to yield the total polyphenolic content. All signals belonging to the same aromatic ring structure in the 2D spectra were averaged, rather than summed, prior to the quantification calculation so as to not yield overestimations. Quantification using Equation 1 is dependent on an unknown factor, namely the number of protons per aromatic ring in the samples (n sample). Increasing this value will decrease the molar concentration calculated; however, using 13C NMR to estimate this value provides a more accurate quantification. 44 Furthermore, a standard molecular weight is required to report the quantification in mass units (mg/g). This value is directly proportional to the quantification result and has a significant impact on the quantification; a high molecular weight standard will yield a higher quantification result.
FIGURE 4.
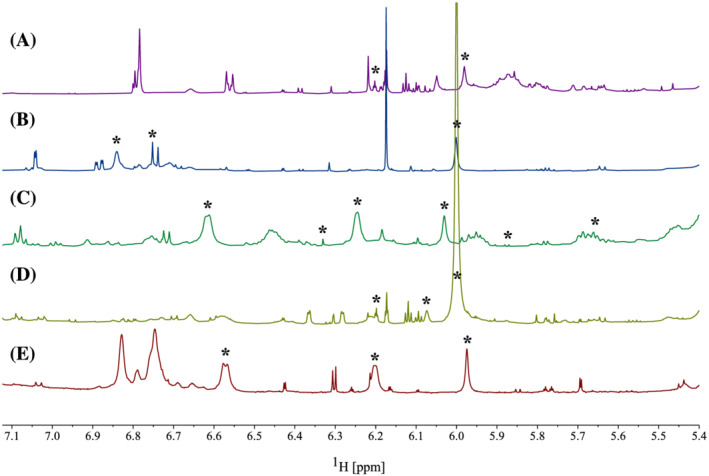
1H NMR spectra displaying the polyphenolic region (7.0–5.5 ppm) used for quantification of Fucus vesiculosus (A), Ascophyllum nodosum (B), Saccharina latissima (C), Laminaria digitata (D), and Laminaria hyperborea M20 (E). Signals labelled with asterisk (*) were deselected based on 2D NMR prior to quantification.
FIGURE 3.
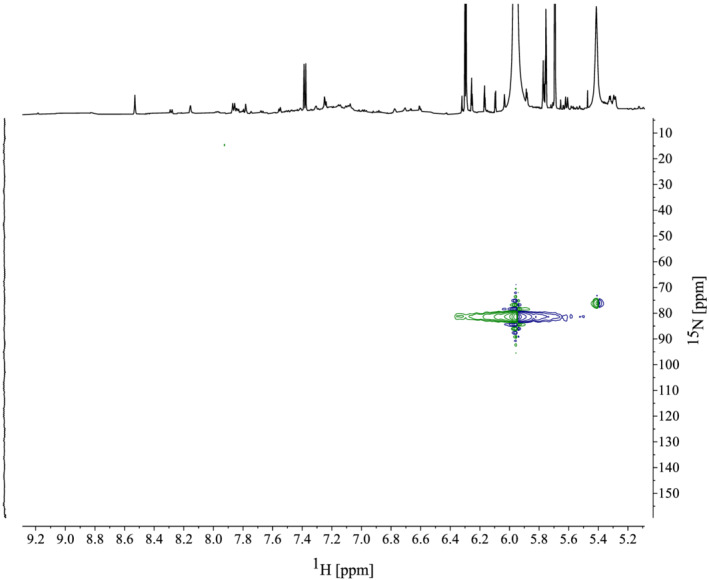
1H‐15N heteronuclear single‐quantum coherence (HSQC) spectrum of Laminaria hyperborea M20 indicating a large peak at 5.97 ppm coupling to a nitrogen at 81.8 ppm, indicating this signal does not represent a polyphenol. Similar peaks observed at 6 ppm in other algae analysed were also eliminated prior to quantification.
The FC TPC assay was also used for quantification. This is a colorimetric assay dependent on the redox reaction of the FC reagent with hydroxyl groups of polyphenols in a sample. Singleton and Rossi's TPC method from 1965, optimised for wine samples, is one of the most cited. 78 Slight modifications of this method were made prior to the analysis to optimise the assay for seaweed samples.
In Table 1, selected reported quantifications of polyphenols from brown seaweeds utilising the TPC assay and/or qNMR are shown. 36 , 44 , 45 , 53 The majority of the works quantify using only the TPC assay; however, some studies use both methods such as Parys et al. (2007), Parys et al. (2009), Vissers et al. (2012), and Ford et al. (2020). 44 , 45 , 53 , 60 Comparison and evaluation of analytical methods for total quantification of polyphenols in seaweeds is relevant to gain new knowledge of the polyphenolic content in seaweeds and to search for the optimal method to assess the total content of this highly diverse group of compounds. 9 , 36 , 53 , 60 However, some studies report results from the TPC and qNMR as different standard equivalents or without any explanation of the standard used for quantification. This makes both interpretation and comparison difficult and highlights the limitation of not having standardised methods.
The quantifications presented using a selective qNMR method resulted in the highest polyphenol content found for S. latissima (1.2% phloroglucinol equivalents (PGE), dry weight (DW)) (Table 5), while the two other Laminariales were found to contain the lowest observed polyphenolic content in the study with 0.41– 0.62% (PGE, DW). Fucus vesiculosus and A. nodosum showed similar values to S. latissima, with polyphenol contents of 1.1% and 0.9% (PGE, DW), respectively. The qNMR results calculated using phloroglucinol (MW = 126.11 g/mol) were slightly lower than once calculated using gallic acid (MW = 170.12 g/mol) due to the lower molecular weight of phloroglucinol, as previously discussed.
Parys et al. (2007) and Ford et al. (2020) both analysed A. nodosum (Fucaceae) and found the total polyphenol content, using qNMR, to be 25.34% trimesic acid equivalents (TAE) and 37.35 mg TAE/g, respectively (Table 1). 45 , 53 These results are considerably higher than the amounts found for the same species in our investigation (Table 5). Whether the reported data by Parys et al and Ford et al are calculated based on fresh or dry weight is not clear, although dry weight concentrations are most frequently used. Additionally, both studies perform NMR quantification using a larger molecular weight for standardisation (MW (trimesic acid) = 210.14 g/mol) and a smaller number of protons (n sample = 1.7). 45 , 53 These two parameters will make a significant impact on the quantification as mentioned previously and therefore contribute to the higher quantification reported by Ford et al More comparable to our results are studies performed by Jégou et al. (2015) reporting 0.46% polyphenol content in Cystoseira tamariscifolia (Fucaceae) using qNMR, and Roleda et al. (2019) reporting TPC amounts of 5–15 mg PGE/g DW in S. latissima, with the latter being season dependent (Table 1). 63 , 65
Applying the TPC assay to the three Laminariales resulted in polyphenolic contents of 0.61%–1.5% (PGE, DW) and 0.54%–1.3% (GAE, DW), with the greatest amounts observed for sugar kelp (S. latissima) (Table 6). Fucus vesiculosus and A. nodosum showed TPC results of approximately 4% and 2% PGE, respectively. Based on the TPC results only, the eulittoral F. vesiculosus and A. nodosum, growing in more shallow waters, show a higher polyphenolic concentration compared with all three Laminariaceae species growing in the sublittoral zone. Parys et al. (2007) and Zhang et al. (2006) present even higher TPC values for species belonging to the Fucaceae family (15.9% PGE and 23.2% PGE, respectively), while Farvin and Jackobsen (2013), O'Sullivan et al. (2011), and Heffernan et al. (2014) all have found lower TPC values for Fucus species (Table 1). 52 , 53 , 54 , 55 , 57 A study of Icelandic seaweeds by Wang et al. (2009) reports higher total polyphenol contents for both F. vesiculosus (⁓ 24%) and L. hyperborea (⁓ 13%). However, their results for L. digitata are comparable to our investigation (10 mg PGE/g, ⁓ 1%). 56
TABLE 6.
Overview of the total polyphenol content obtained for Laminaria hyperborea, Laminaria digitata, Saccharina latissimia, Ascophyllum nodosum, and Fucus vesiculosus extracts using the optimised total phenolic content (TPC) reaction conditions. Results are expressed as gallic acid and phloroglucinol equivalents (GAE/PGE) per dry weight (DW) (mean ± SD, n = 3).
| Seaweed | Family | Zone | C [mg GAE/g DW] | C [mg PGE/g DW] |
|---|---|---|---|---|
| L. hyperborea M20 | Laminariaceae | Sublittoral | 6.23 ± 0.11 | 7.15 ± 0.15 |
| L. hyperborea S20 | Laminariaceae | Sublittoral | 5.72 ± 0.07 | 6.56 ± 0.08 |
| L. hyperborea A21 | Laminariaceae | Sublittoral | 5.35 ± 0.04 | 6.14 ± 0.05 |
| L. digitata | Laminariaceae | Sublittoral | 6.94 ± 0.09 | 7.93 ± 0.09 |
| S. latissima | Laminariaceae | Sublittoral | 13.1 ± 0.04 | 15.0 ± 0.05 |
| A. nodosum | Fucaceae | Eulittoral | 17.6 ± 0.04 | 20.1 ± 0.05 |
| F. vesiculosus | Fucaceae | Eulittoral | 37.0 ± 1.0 | 42.0 ± 1.1 |
3.3. Comparing the selective qNMR method with the TPC assay
In Figure 5 the quantified polyphenolic content of L. hyperborea, harvested in various seasons, L. digitata, S. latissima, A. nodosum, and F. vesiculosus using the selective qNMR method and the TPC assay are shown—including a trendline for the two methods compared with a non‐selective qNMR method. The general trend shows higher TPC values compared with the corresponding qNMR quantifications. Minor differences were observed between the two methods for the sublittoral growing Laminariaceae species (L. hyperborea, L. digitata, and S. latissima) (Figure 5). Indications of possible seasonal differences were observed for the selected samples, although a complete seasonal study was not performed. However, variations of the polyphenol content in regard to harvest season have previously been reported in literature. 9 , 45 , 60 , 65 , 90 A significant difference between the TPC and qNMR results was observed for both eulittoral growing Fucaceae species A. nodosum and F. vesiculosus, where the TPC assay yields up to three time the amount found with the qNMR method for the Fucus species. Ford et al. (2020) also reports higher TPC values compared with their qNMR results for a Fucaceae species F. serratus. 45 However, they use a non‐selective qNMR method, and their qNMR quantification of A. nodosum is approximately 20% higher compared with their TPC assay. Parys et al. (2009) also compare the FC TPC assay with a (non‐selective) qNMR method in a seasonal investigation of the polyphenol concentration in A. nodosum. 60 Their FC TPC assay yields 1.5–4 times higher polyphenolic amounts than their qNMR method, and Parys et al. conclude that the results from the two methods cannot be compared due to their principal differences. Both Ford et al. and Parys et al. apply a non‐selective qNMR method, and these results are higher than the ones presented in our study. A significant difference can be observed between selective and non‐selective qNMR quantification, as illustrated in Figure 5.
FIGURE 5.
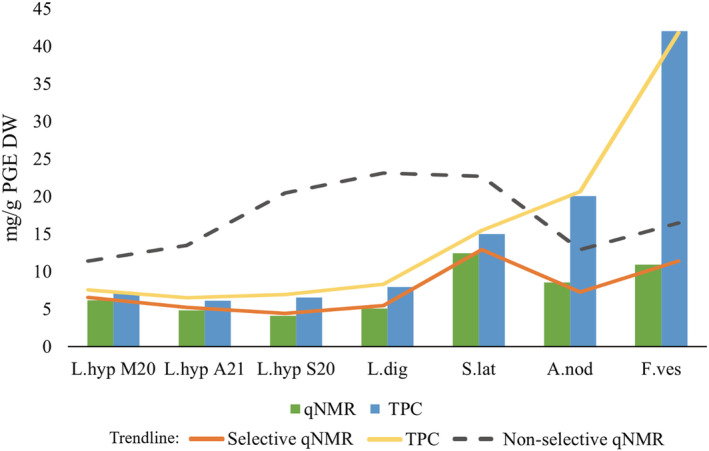
Polyphenolic content of Laminaria hyperborea (L.hyp) (harvested in various seasons M20 = March 2020, A21 = August 2021, S20 = September 2020), Laminaria digitata (L.dig), Saccharina latissima, Ascophyllum nodosum, and Fucus vesiculosus quantified using qNMR (green) and total phenolic content (TPC) assay (blue), reported as milligram phloroglucinol equivalents (PGE) per gram dry weight (DW). The orange and yellow lines represent the trend lines of the displayed bar chart, and the grey line indicates non‐selective qNMR values.
In general, the colorimetric TPC assay using the FC reagent has been assumed to overestimate the polyphenolic content. 69 , 70 , 91 , 92 This is due to several factors, such as the presence of metal contaminants or high levels of reducing sugars or other compounds, for instance ascorbic acid or amino acids, which interfere with the FC reaction. 68 , 91 , 93 However, increased polyphenol diversity within the extract, such as hydroxybenzoic acids, hydroxycinnamic acids, hydrolysable tannins, proanthocyanins, and flavonoids—seen in seaweed species in rather shallow waters as well as in aquatic and terrestrial plants—may also result in higher TPC quantifications due to the possibly larger number of reacting groups within a molecule not accounted for in the TPC standardisation. 9 , 36 , 75 , 77 , 78 , 90 , 94 , 95 , 96 Quantitative NMR as a method is in general not as sensitive as the colorimetric TPC assay to interfering species such as metal contaminants, high levels of reducing sugars, or pigments. The reported selective qNMR method is also less influenced by the diversity of the polyphenols, as the 13C NMR partial characterisation prior to the quantification facilitates the estimate of number of protons (H) per aromatic ring of the dominating polyphenol group (phlorotannins) in the extract—increasing the accuracy of the method.
The polyphenolic content will always reflect the variety of biosynthesis' found within different species and external factors such as temperature, UV exposure, pathogens, etc. that will always vary within habitats, sites, and seasons, influencing both the polyphenolic production and the production of other metabolites—the latter particularly affecting the non‐selective colorimetric TPC quantification. Results reported herein reveal that the TPC method can possibly be safely applied to sublittoral growing Laminariales species, which most likely possess a less diverse polyphenolic content and fewer interfering species. However, for the shallower‐growing seaweed species, such as the eulittoral F. vesiculosus and A. nodosum, the TPC assay and the qNMR method show significant differences, most likely reflecting the shortcomings of the colorimetric assay. By applying a selective qNMR method for total polyphenolic quantification, the results will be less influenced by the diversity of the polyphenols in the sample and the presence of interfering compounds than when using the TPC assay. Hence, this approach will provide a polyphenolic quantification assumed to be closer to the “true” polyphenol concentration of brown seaweeds.
ACKNOWLEDGMENTS
M.E.W. gratefully acknowledges the Norwegian Research Council (project NFR297507) and Alginor ASA for their fellowship and Georg Kopplin for guidance. The authors acknowledge Jarl Underhaug for great guidance and support when performing NMR experiments at the Norwegian NMR Platform (NNP). The authors acknowledge the Ocean Forest project at Lerøy AS for providing Saccharina latissima material. This work was partly supported by the Bergen Research Foundation (BFS‐NMR‐1), Sparebankstiftinga Sogn og Fjordane (509‐42/16), and the Research Council of Norway through the Norwegian NMR Platform, NNP (226244/F50).
Wekre ME, Hellesen Brunvoll S, Jordheim M. Advancing quantification methods for polyphenols in brown seaweeds—applying a selective qNMR method compared with the TPC assay. Phytochemical Analysis. 2022;33(7):1099‐1110. doi: 10.1002/pca.3162
Funding information Bergen Research Foundation, Grant/Award Number: BFS‐NMR‐1; Norges Forskningsråd, Grant/Award Number: 297507; Sparebankstiftinga Sogn og Fjordane, Grant/Award Number: 509‐42/16; Research Council of Norway through the Norwegian NMR Platform, NNP, Grant/Award Number: 226244/F50
DATA AVAILABILITY STATEMENT
The supplementary information is deposited in UoB Open Research Data (https://doi.org/10.18710/VZHSWT). The deposited data contain information on the TPC assay and qNMR results.
REFERENCES
- 1. Langton R, Augyte S, Price N, et al. An Ecosystem Approach to the Culture of Seaweed. 24 (2019). [Google Scholar]
- 2. Collins JE, Vanagt T, Huys I, Vieira H. Marine Bioresource Development – Stakeholder's Challenges, Implementable Actions, and Business Models. Front Mar Sci. 2020;7:62. doi: 10.3389/fmars.2020.00062 [DOI] [Google Scholar]
- 3. Stévant P, Rebours C, Chapman A. Seaweed aquaculture in Norway: recent industrial developments and future perspectives. Aquac Int. 2017;25:1373‐1390. [Google Scholar]
- 4. Holdt SL, Kraan S. Bioactive compounds in seaweed: functional food applications and legislation. J Appl Phycol. 2011;23:543‐597. [Google Scholar]
- 5. Domínguez H. Algae as a source of biologically active ingredients for the formulation of functional foods and nutraceuticals. In: Functional Ingredients from Algae for Foods and Nutraceuticals. Elsevier; 2013:1‐19. doi: 10.1533/9780857098689.1 [DOI] [Google Scholar]
- 6. Mahadevan K. Seaweeds: a sustainable food source. In: Seaweed Sustainability. Elsevier; 2015:347‐364. doi: 10.1016/B978-0-12-418697-2.00013-1 [DOI] [Google Scholar]
- 7. Mahalik NP, Kim K. Aquaculture Monitoring and Control Systems for Seaweed and Fish Farming. World J Agric Res. 2014;2:176‐182. [Google Scholar]
- 8. Ktari L, Chebil Ajjabi L, De Clerck O, Gómez Pinchetti JL, Rebours C. Seaweeds as a promising resource for blue economy development in Tunisia: current state, opportunities, and challenges. J Appl Phycol. 2021;34(1):489‐505. doi: 10.1007/s10811-021-02579-w [DOI] [Google Scholar]
- 9. Ford L, Theodoridou K, Sheldrake GN, Walsh PJ. A critical review of analytical methods used for the chemical characterisation and quantification of phlorotannin compounds in brown seaweeds. Phytochem Anal. 2019;30:587‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hudek K, Davis LC, Ibbini J, Erickson L. Commercial Products from Algae. In: Bajpai R, Prokop A, Zappi M, eds. Algal Biorefineries. Springer Netherlands; 2014:275‐295. doi: 10.1007/978-94-007-7494-0_11 [DOI] [Google Scholar]
- 11. Boukid F, Castellari M. Food and Beverages Containing Algae and Derived Ingredients Launched in the Market from 2015 to 2019: A Front‐of‐Pack Labeling Perspective with a Special Focus on Spain. Foods. 2021;10(1):173. doi: 10.3390/foods10010173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murray M, Dordevic AL, Ryan L, Bonham MP. An emerging trend in functional foods for the prevention of cardiovascular disease and diabetes: Marine algal polyphenols. Crit Rev Food Sci Nutr. 2018;58:1342‐1358. [DOI] [PubMed] [Google Scholar]
- 13. Cajnko MM, Novak U, Likozar B. Cascade valorization process of brown alga seaweed Laminaria hyperborea by isolation of polyphenols and alginate. J Appl Phycol. 2019;31:3915‐3924. [Google Scholar]
- 14. Goleniowski M, Bonfill M, Cusido R, Palazón J. Phenolic Acids. In: Ramawat KG, Mérillon J‐M, eds. Natural Products. Springer Berlin Heidelberg; 2013:1951‐1973. doi: 10.1007/978-3-642-22144-6_64 [DOI] [Google Scholar]
- 15. Jiménez‐Escrig A, Jiménez‐Jiménez I, Pulido R, Saura‐Calixto F. Antioxidant activity of fresh and processed edible seaweeds: Antioxidant activity of seaweeds. J Sci Food Agric. 2001;81:530‐534. [Google Scholar]
- 16. Matsukawa R, Dubinsky Z, Kishimoto E, et al. A comparison of screening methods for antioxidant activity in seaweeds. J Appl Phycol. 1997;9(1):29‐35. doi: 10.1023/A:1007935218120 [DOI] [Google Scholar]
- 17. Nasseri MA, Behravesh S, Allahresani A, Kazemnejadi M. Phytochemical and antioxidant studies of Cleome heratensis (Capparaceae) plant extracts. Bioresour Bioprocess. 2019;6(1):5. doi: 10.1186/s40643-019-0240-1 [DOI] [Google Scholar]
- 18. Cotas J, Leandro A, Monteiro P, et al. Seaweed Phenolics: From Extraction to Applications. Mar Drugs. 2020;18(8):384. doi: 10.3390/md18080384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Svensson CJ, Pavia H, Toth GB. Do plant density, nutrient availability, and herbivore grazing interact to affect phlorotannin plasticity in the brown seaweed Ascophyllum nodosum. Mar Biol. 2007;151:2177‐2181. [Google Scholar]
- 20. Pavia H, Cervin G, Lindgren A, Åberg P. Effects of UV‐B radiation and simulated herbivory on phlorotannins in the brown alga Ascophyllum nodosum. Mar Ecol Prog Ser. 1997;157:139‐146. [Google Scholar]
- 21. Rönnberg O, Ruokolahti C. Seasonal variation of algal epiphytes and phenolic content of Fucus vesiculosus in a northern Baltic archipelago. Ann Bot Fenn. 1986;23:317‐323. [Google Scholar]
- 22. Pangestuti R, Kim S‐K. Biological activities and health benefit effects of natural pigments derived from marine algae. J Funct Foods. 2011;3:255‐266. [Google Scholar]
- 23. Li Y‐X, Wijesekara I, Li Y, Kim S‐K. Phlorotannins as bioactive agents from brown algae. Process Biochem. 2011;46:2219‐2224. [Google Scholar]
- 24. Heo S, Park E, Lee K, Jeon Y. Antioxidant activities of enzymatic extracts from brown seaweeds. Bioresour Technol. 2005;96:1613‐1623. [DOI] [PubMed] [Google Scholar]
- 25. Wijesekara I, Yoon NY, Kim S‐K. Phlorotannins from Ecklonia cava (Phaeophyceae): Biological activities and potential health benefits. Biofactors. 2010;36(6):408‐414. doi: 10.1002/biof.114 [DOI] [PubMed] [Google Scholar]
- 26. Besednova NN, Andryukov BG, Zaporozhets TS, et al. Antiviral Effects of Polyphenols from Marine Algae. Biomedicine. 2021;9(2):200. doi: 10.3390/biomedicines9020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwon H‐J, Ryu YB, Kim YM, et al. In vitro antiviral activity of phlorotannins isolated from Ecklonia cava against porcine epidemic diarrhea coronavirus infection and hemagglutination. Bioorg Med Chem. 2013;21(15):4706‐4713. doi: 10.1016/j.bmc.2013.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Catarino MD, Amarante SJ, Mateus N, Silva AMS, Cardoso SM. Brown Algae Phlorotannins: A Marine Alternative to Break the Oxidative Stress, Inflammation and Cancer Network. Foods. 2021;10(7):1478. doi: 10.3390/foods10071478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahn M‐J, Yoon KD, Min SY, et al. Inhibition of HIV‐1 Reverse Transcriptase and Protease by Phlorotannins from the Brown Alga Ecklonia cava. Biol Pharm Bull. 2004;27(4):544‐547. doi: 10.1248/bpb.27.544 [DOI] [PubMed] [Google Scholar]
- 30. Artan M, Li Y, Karadeniz F, Lee SH, Kim MM, Kim SK. Anti‐HIV‐1 activity of phloroglucinol derivative, 6,6′‐bieckol, from Ecklonia cava. Bioorg Med Chem. 2008;16(17):7921‐7926. doi: 10.1016/j.bmc.2008.07.078 [DOI] [PubMed] [Google Scholar]
- 31. Kong C‐S, Kim J‐A, Yoon N‐Y, Kim S‐K. Induction of apoptosis by phloroglucinol derivative from Ecklonia Cava in MCF‐7 human breast cancer cells. Food Chem Toxicol. 2009;47:1653‐1658. [DOI] [PubMed] [Google Scholar]
- 32. Park J‐Y, Kim JH, Kwon JM, et al. Dieckol, a SARS‐CoV 3CLpro inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg Med Chem. 2013;21(13):3730‐3737. doi: 10.1016/j.bmc.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ganesan AR, Tiwari U, Rajauria G. Seaweed nutraceuticals and their therapeutic role in disease prevention. Food Sci Human Wellness. 2019;8:252‐263. [Google Scholar]
- 34. Zhou N, Gu X, Zhuang T, Xu Y, Yang L, Zhou M. Gut Microbiota: A Pivotal Hub for Polyphenols as Antidepressants. J Agric Food Chem. 2020;68(22):6007‐6020. doi: 10.1021/acs.jafc.0c01461 [DOI] [PubMed] [Google Scholar]
- 35. Tong T, Liu X, Yu C. Extraction and Nano‐Sized Delivery Systems for Phlorotannins to Improve Its Bioavailability and Bioactivity. Mar Drugs. 2021;19(11):625. doi: 10.3390/md19110625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wekre ME, Kåsin K, Underhaug J, Holmelid B, Jordheim M. Quantification of Polyphenols in Seaweeds: A Case Study of Ulva intestinalis. Antioxidants. 2019;8(12):612 doi: 10.3390/antiox8120612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zenthoefer M, Geisen U, Hofmann‐Peiker K, et al. Isolation of polyphenols with anticancer activity from the Baltic Sea brown seaweed Fucus vesiculosus using bioassay‐guided fractionation. J Appl Phycol. 2017;29(4):2021‐2037. doi: 10.1007/s10811-017-1080-z [DOI] [Google Scholar]
- 38. Koivikko R, Loponen J, Honkanen T, Jormalainen V. CONTENTS OF SOLUBLE, CELL‐WALL‐BOUND AND EXUDED PHLOROTANNINS IN THE BROWN ALGA Fucus vesiculosus, WITH IMPLICATIONS ON THEIR ECOLOGICAL FUNCTIONS. J Chem Ecol. 2005;31(1):195‐212. doi: 10.1007/s10886-005-0984-2 [DOI] [PubMed] [Google Scholar]
- 39. Steevensz AJ, MacKinnon SL, Hankinson R, et al. Profiling Phlorotannins in Brown Macroalgae by Liquid Chromatography‐High Resolution Mass Spectrometry: Phlorotannins in Seaweed by LC‐HRMS. Phytochem Anal. 2012;23(5):547‐553. doi: 10.1002/pca.2354 [DOI] [PubMed] [Google Scholar]
- 40. Mwangi HM, Njue WM, Onani MO, Thovhoghi N, Mabusela WT. Phlorotannins and a sterol isolated from a brown alga Ecklonia maxima, and their cytotoxic activity against selected cancer cell lines HeLa, H157 and MCF7. Interdiscip J Chem. 2017;2(2):1‐6. doi: 10.15761/IJC.1000120 [DOI] [Google Scholar]
- 41. Sardari RRR, Prothmann J, Gregersen O, Turner C, Nordberg Karlsson E. Identification of Phlorotannins in the Brown Algae, Saccharina latissima and Ascophyllum nodosum by Ultra‐High‐Performance Liquid Chromatography Coupled to High‐Resolution Tandem Mass Spectrometry. Molecules. 2020;26(1):43. doi: 10.3390/molecules26010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fernando IPS, Lee W, Ahn G. Marine algal flavonoids and phlorotannins; an intriguing frontier of biofunctional secondary metabolites. Crit Rev Biotechnol. 2022;42(1):23‐45. doi: 10.1080/07388551.2021.1922351 [DOI] [PubMed] [Google Scholar]
- 43. Singh IP, Sidana J. Phlorotannins. In: Functional Ingredients from Algae for Foods and Nutraceuticals. Elsevier; 2013:181‐204. doi: 10.1533/9780857098689.1.181 [DOI] [Google Scholar]
- 44. Vissers AM, Caligiani A, Sforza S, Vincken J‐P, Gruppen H. Phlorotannin Composition of Laminaria digitata: Phlorotannin composition of Laminaria digitata. Phytochem Anal. 2017;28:487‐495. [DOI] [PubMed] [Google Scholar]
- 45. Ford L, Stratakos AC, Theodoridou K, et al. Polyphenols from Brown Seaweeds as a Potential Antimicrobial Agent in Animal Feeds. ACS Omega. 2020;5(16):9093‐9103. doi: 10.1021/acsomega.9b03687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Monbet P, Worsfold P, McKelvie I. Advances in marine analytical chemistry. Talanta. 2019;202:610. doi: 10.1016/j.talanta.2019.03.097 [DOI] [Google Scholar]
- 47. Wang Z‐F, You Y‐L, Li F‐F, Kong W‐R, Wang S‐Q. Research Progress of NMR in Natural Product Quantification. Molecules. 2021;26(20):6308. doi: 10.3390/molecules26206308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferdouse F, Holdt SL, Smith R, Murúa P, Yang Z. The global status of seaweed production, trade and utilizationGLOBEFISH Res. Programme. Vol. 124I; 2018. [Google Scholar]
- 49. Murray M, Dordevic AL, Ryan L, Bonham MP. Phlorotannins and Macroalgal Polyphenols: Potential As Functional Food Ingredients and Role in Health Promotion. In: Rani V, Yadav UC, eds. Functional Food and Human Health. Springer Singapore; 2018:27‐58. [Google Scholar]
- 50. Williamson G, Manach C. Bioavailability and bioefficacy of polyphenols in humans. II. Review of 93 intervention studies. Am J Clin Nutr. 2005;81(1):243S‐255S. doi: 10.1093/ajcn/81.1.243S [DOI] [PubMed] [Google Scholar]
- 51. Stern JL, Hagerman AE, Steinberg PD, Winter FC, Estes JA. A new assay for quantifying brown algal phlorotannins and comparisons to previous methods. J Chem Ecol. 1996;22:1273‐1293. [DOI] [PubMed] [Google Scholar]
- 52. Farvin KHS, Jacobsen C. Phenolic compounds and antioxidant activities of selected species of seaweeds from Danish coast. Food Chem. 2013;138:1670‐1681. [DOI] [PubMed] [Google Scholar]
- 53. Parys S, Rosenbaum A, Kehraus S, Reher G, Glombitza KW, König GM. Evaluation of Quantitative Methods for the Determination of Polyphenols in Algal Extracts. J Nat Prod. 2007;70(12):1865‐1870. doi: 10.1021/np070302f [DOI] [PubMed] [Google Scholar]
- 54. O'Sullivan AM, O'Callaghan YC, O'Grady MN, et al. In vitro and cellular antioxidant activities of seaweed extracts prepared from five brown seaweeds harvested in spring from the west coast of Ireland. Food Chem. 2011;126(3):1064‐1070. doi: 10.1016/j.foodchem.2010.11.127 [DOI] [Google Scholar]
- 55. Zhang Q, Zhang J, Shen J, Silva A, Dennis DA, Barrow CJ. A Simple 96‐Well Microplate Method for Estimation of Total Polyphenol Content in Seaweeds. J Appl Phycol. 2006;18(3–5):445‐450. doi: 10.1007/s10811-006-9048-4 [DOI] [Google Scholar]
- 56. Wang T, Jónsdóttir R, Ólafsdóttir G. Total phenolic compounds, radical scavenging and metal chelation of extracts from Icelandic seaweeds. Food Chem. 2009;116:240‐248. [Google Scholar]
- 57. Heffernan N, Smyth TJ, FitzGerald RJ, Soler‐Vila A, Brunton N. Antioxidant activity and phenolic content of pressurised liquid and solid‐liquid extracts from four Irish origin macroalgae. Int J Food Sci Technol. 2014;49:1765‐1772. [Google Scholar]
- 58. Tierney MS, Smyth TJ, Hayes M, Soler‐Vila A, Croft AK, Brunton N. Influence of pressurised liquid extraction and solid‐liquid extraction methods on the phenolic content and antioxidant activities of Irish macroalgae. Int J Food Sci Technol. 2013;48(4):860‐869. doi: 10.1111/ijfs.12038 [DOI] [Google Scholar]
- 59. Gisbert M, Barcala M, Rosell CM, Sineiro J, Moreira R. Aqueous extracts characteristics obtained by ultrasound‐assisted extraction from Ascophyllum nodosum seaweeds: effect of operation conditions. J Appl Phycol. 2021;33(5):3297‐3308. doi: 10.1007/s10811-021-02546-5 [DOI] [Google Scholar]
- 60. Parys S, Kehraus S, Pete R, Küpper FC, Glombitza KW, König GM. Seasonal variation of polyphenolics in Ascophyllum nodosum (Phaeophyceae). Eur J Phycol. 2009;44(3):331‐338. doi: 10.1080/09670260802578542 [DOI] [Google Scholar]
- 61. Anaëlle T, Serrano Leon E, Laurent V, et al. Green improved processes to extract bioactive phenolic compounds from brown macroalgae using Sargassum muticum as model. Talanta. 2013;104:44‐52. doi: 10.1016/j.talanta.2012.10.088 [DOI] [PubMed] [Google Scholar]
- 62. Li Y, Fu X, Duan D, Liu X, Xu J, Gao X. Extraction and Identification of Phlorotannins from the Brown Alga, Sargassum fusiforme (Harvey) Setchell. Mar Drugs. 2017;15(2):49. doi: 10.3390/md15020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jégou C, Kervarec N, Cérantola S, Bihannic I, Stiger‐Pouvreau V. NMR use to quantify phlorotannins: The case of Cystoseira tamariscifolia, a phloroglucinol‐producing brown macroalga in Brittany (France). Talanta. 2015;135:1‐6. doi: 10.1016/j.talanta.2014.11.059 [DOI] [PubMed] [Google Scholar]
- 64. Leyton A, Pezoa‐Conte R, Barriga A, et al. Identification and efficient extraction method of phlorotannins from the brown seaweed Macrocystis pyrifera using an orthogonal experimental design. Algal Res. 2016;16:201‐208. doi: 10.1016/j.algal.2016.03.019 [DOI] [Google Scholar]
- 65. Roleda MY, Marfaing H, Desnica N, et al. Variations in polyphenol and heavy metal contents of wild‐harvested and cultivated seaweed bulk biomass: Health risk assessment and implication for food applications. Food Control. 2019;95:121‐134. doi: 10.1016/j.foodcont.2018.07.031 [DOI] [Google Scholar]
- 66. Folin O, Ciocalteu V. ON TYROSINE AND TRYPTOPHANE DETERMINATIONS IN PROTEINS. J Biol Chem. 1927;73(2):627‐650. doi: 10.1016/S0021-9258(18)84277-6 [DOI] [Google Scholar]
- 67. Ikawa M, Schaper TD, Dollard CA, Sasner JJ. Utilization of Folin−Ciocalteu Phenol Reagent for the Detection of Certain Nitrogen Compounds. J Agric Food Chem. 2003;51:1811‐1815. [DOI] [PubMed] [Google Scholar]
- 68. Jacobsen C, Sørensen A‐DM, Holdt SL, Akoh CC, Hermund DB. Source, Extraction, Characterization, and Applications of Novel Antioxidants from Seaweed. Annu Rev Food Sci Technol. 2019;10:541‐568. [DOI] [PubMed] [Google Scholar]
- 69. van Alstyne KL. Comparison of three methods for quantifying brown algal polyphenolic compounds. J Chem Ecol. 1995;21:45‐58. [DOI] [PubMed] [Google Scholar]
- 70. Pękal A, Pyrzynska K. Evaluation of Aluminium Complexation Reaction for Flavonoid Content Assay. Food Anal Methods. 2014;7:1776‐1782. [Google Scholar]
- 71. Jordheim M, Aaby K, Fossen T, Skrede G, Andersen ØM. Molar Absorptivities and Reducing Capacity of Pyranoanthocyanins and Other Anthocyanins. J Agric Food Chem. 2007;55:10591‐10598. [DOI] [PubMed] [Google Scholar]
- 72. Pauli GF, Gödecke T, Jaki BU, Lankin DC. Quantitative 1 H NMR. Development and Potential of an Analytical Method: An Update. J Nat Prod. 2012;75:834‐851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pauli GF, Jaki BU, Lankin DC. Quantitative1H NMR: Development and Potential of a Method for Natural Products Analysis. J Nat Prod. 2005;68(1):133‐149. doi: 10.1021/np0497301 [DOI] [PubMed] [Google Scholar]
- 74. Nerantzaki AA, Tsiafoulis CG, Charisiadis P, Kontogianni VG, Gerothanassis IP. Novel determination of the total phenolic content in crude plant extracts by the use of 1H NMR of the –OH spectral region. Anal Chim Acta. 2011;688:54‐60. doi: 10.1016/j.aca.2010.12.027 [DOI] [PubMed] [Google Scholar]
- 75. Sævdal Dybsland C, Bekkby T, Hasle Enerstvedt K, Kvalheim OM, Rinde E, Jordheim M. Variation in Phenolic Chemistry in Zostera marina Seagrass along Environmental Gradients. Plan Theory. 2021;10(2):334. doi: 10.3390/plants10020334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ummat V, Tiwari BK, Jaiswal AK, et al. Optimisation of Ultrasound Frequency, Extraction Time and Solvent for the Recovery of Polyphenols, Phlorotannins and Associated Antioxidant Activity from Brown Seaweeds. Mar Drugs. 2020;18(5):250 doi: 10.3390/md18050250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Singleton VL, Orthofer R, Lamuela‐Raventós RM. [14] Analysis of total phenols and other oxidation substrates and antioxidants by means of folin‐ciocalteu reagent. In: Methods in Enzymology, Vol. 299. Elsevier; 1999:152‐178. doi: 10.1016/S0076-6879(99)99017-1 [DOI] [Google Scholar]
- 78. Singleton VL, Rossi JA. Colorimetry of Total Phenolics with Phosphomolybdic‐Phosphotungstic Acid Reagents. Am J Enol Vitic. 1965;16:144. [Google Scholar]
- 79. Ün İ, Ün ŞŞ, Tanrıkulu N, Ünlü A, Ok S. Assessing the concentration of conjugated fatty acids within pomegranate seed oil using quantitative nuclear magnetic resonance (qNMR). Phytochem Anal. 2021;33:452‐459. doi: 10.1002/pca.3101 [DOI] [PubMed] [Google Scholar]
- 80. Piotto M, Bourdonneau M, Elbayed K, Wieruszeski J‐M, Lippens G. New DEFT sequences for the acquisition of one‐dimensional carbon NMR spectra of small unlabelled molecules. Magn Reson Chem. 2006;44:943‐947. [DOI] [PubMed] [Google Scholar]
- 81. Zhang K, Tieke B, Forgie JC, Vilela F, Parkinson JA, Skabara PJ. Cross‐linked polymers based on 2,3,5,6‐tetra‐substituted pyrrolo[3,4‐c]pyrrole‐1,4(2H,5H)‐dione (DPP): Synthesis, optical and electronic properties. Polymer. 2010;51(26):6107‐6114. doi: 10.1016/j.polymer.2010.10.054 [DOI] [Google Scholar]
- 82. Pavia H, Toth GB. Influence of light and nitrogen on the phlorotannin content of the brown seaweeds Ascophyllum nodosum and Fucus vesiculosus. Hydrobiologia. 2000;440(1/3):299‐305. doi: 10.1023/A:1004152001370 [DOI] [Google Scholar]
- 83. Tabassum MR, Xia A, Murphy JD. Seasonal variation of chemical composition and biomethane production from the brown seaweed Ascophyllum nodosum. Bioresour Technol. 2016;216:219‐226. doi: 10.1016/j.biortech.2016.05.071 [DOI] [PubMed] [Google Scholar]
- 84. Schiener P, Black KD, Stanley MS, Green DH. The seasonal variation in the chemical composition of the kelp species Laminaria digitata, Laminaria hyperborea, Saccharina latissima and Alaria esculenta. J Appl Phycol. 2015;27(1):363‐373. doi: 10.1007/s10811-014-0327-1 [DOI] [Google Scholar]
- 85. Lopes G, Barbosa M, Vallejo F, et al. Profiling phlorotannins from Fucus spp. of the Northern Portuguese coastline: Chemical approach by HPLC‐DAD‐ESI/MS and UPLC‐ESI‐QTOF/MS. Algal Res. 2018;29:113‐120. doi: 10.1016/j.algal.2017.11.025 [DOI] [Google Scholar]
- 86. Ragan MA, Jamieson WD. Oligomeric polyphloroglucinols from Fucus vesiculosus: Photoplate mass spectrometric investigation. Phytochemistry. 1982;21(11):2709‐2711. doi: 10.1016/0031-9422(82)83103-8 [DOI] [Google Scholar]
- 87. Hermund DB, Plaza M, Turner C, et al. Structure dependent antioxidant capacity of phlorotannins from Icelandic Fucus vesiculosus by UHPLC‐DAD‐ECD‐QTOFMS. Food Chem. 2018;240:904‐909. doi: 10.1016/j.foodchem.2017.08.032 [DOI] [PubMed] [Google Scholar]
- 88. Heffernan N, Brunton NP, FitzGerald RJ, Smyth TJ. Profiling of the Molecular Weight and Structural Isomer Abundance of Macroalgae‐Derived Phlorotannins. Mar Drugs. 2015;13(1):509‐528. doi: 10.3390/md13010509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Montero L, Sánchez‐Camargo AP, García‐Cañas V, et al. Anti‐proliferative activity and chemical characterization by comprehensive two‐dimensional liquid chromatography coupled to mass spectrometry of phlorotannins from the brown macroalga Sargassum muticum collected on North‐Atlantic coasts. J Chromatogr a. 2016;1428:115‐125. doi: 10.1016/j.chroma.2015.07.053 [DOI] [PubMed] [Google Scholar]
- 90. Caro Y, Anamale L, Fouillaud M, Laurent P, Petit T, Dufosse L. Natural hydroxyanthraquinoid pigments as potent food grade colorants: an overview. Nat Prod Bioprospecting. 2012;2(5):174‐193. doi: 10.1007/s13659-012-0086-0 [DOI] [Google Scholar]
- 91. Huang D, Ou B, Prior RL. The Chemistry behind Antioxidant Capacity Assays. J Agric Food Chem. 2005;53(6):1841‐1856. doi: 10.1021/jf030723c [DOI] [PubMed] [Google Scholar]
- 92. Castro‐Alves VC, Cordenunsi BR. Total Soluble Phenolic Compounds Quantification Is Not As Simple As It Seems. Food Anal Methods. 2015;8:873‐884. [Google Scholar]
- 93. Prior RL, Wu X, Schaich K. Standardized Methods for the Determination of Antioxidant Capacity and Phenolics in Foods and Dietary Supplements. J Agric Food Chem. 2005;53:4290‐4302. [DOI] [PubMed] [Google Scholar]
- 94. López‐Hidalgo C, Meijón M, Lamelas L, Valledor L. The rainbow protocol: A sequential method for quantifying pigments, sugars, free amino acids, phenolics, flavonoids and mda from a small amount of sample. Plant Cell Environ. 2021;44(6):1977‐1986. doi: 10.1111/pce.14007 [DOI] [PubMed] [Google Scholar]
- 95. Sanoner P, Guyot S, Marnet N, Molle D, Drilleau J‐F. Polyphenol Profiles of French Cider Apple Varieties (Malus domestica sp.). J Agric Food Chem. 1999;47(12):4847‐4853. doi: 10.1021/jf990563y [DOI] [PubMed] [Google Scholar]
- 96. Wong MC, Griffiths G, Vercaemer B. Seasonal Response and Recovery of Eelgrass (Zostera marina) to Short‐Term Reductions in Light Availability. Estuaries Coast. 2020;43(1):120‐134. doi: 10.1007/s12237-019-00664-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The supplementary information is deposited in UoB Open Research Data (https://doi.org/10.18710/VZHSWT). The deposited data contain information on the TPC assay and qNMR results.


