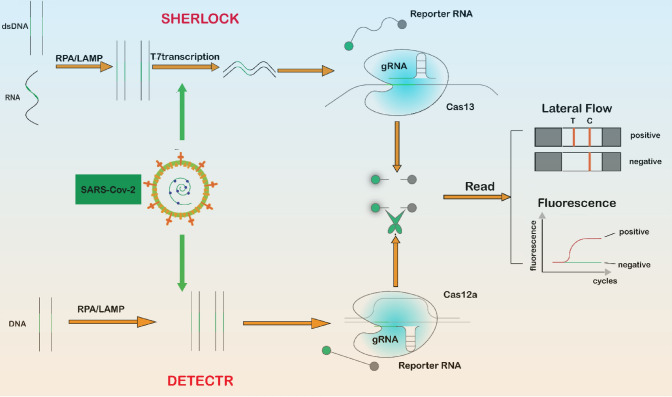
Fig. 4.
CRISPR-based severe acute respiratory syndrome coronavirus 2 detection. Schematic of two CRISPR-based detection methods: (1) SHERLOCK (Specific High Sensitivity Enzymatic Reporter Unlocking): DNA or RNA in the sample is isothermally amplified by recombinase polymerase amplification (RPA) or loop-mediated isothermal amplification (LAMP), the amplicon is converted into RNA by binding to T7 transcriptase, Cas13 is activated when it encounters the target RNA, and the RNA is cut with fluorescent molecules to release fluorescent signals. (2) DETECTR (DNA endonuclease-targeted CRISPR reporter): the target DNA is isothermally amplified by RPA or LAMP, and when Cas12a is activated when it encounters the target DNA and randomly cuts the RNA with fluorescent molecules, releasing a fluorescent signal. dsDNA double-stranded DNA