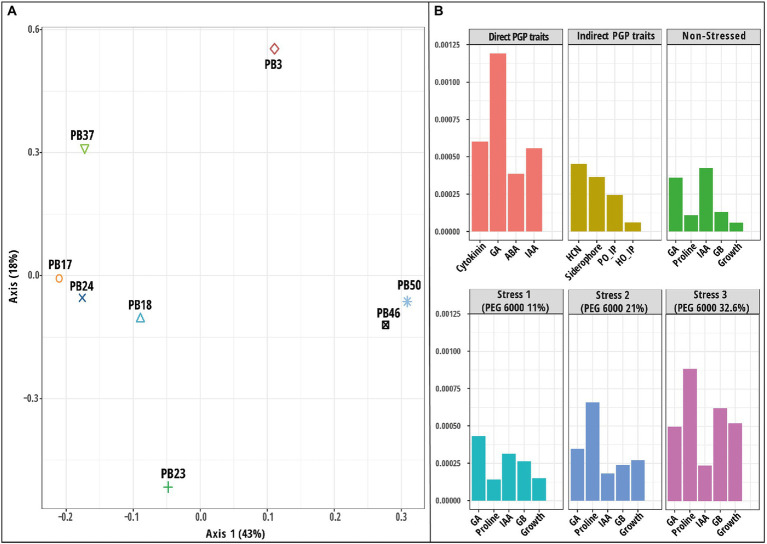
Figure 6.
Results of multiple kernel learning analysis (MKL). (A) A plot of kernel principal component analysis (KPCA) based on six datasets (direct plant growth-promoting (PGP) traits, indirect PGP traits, microbial parameters under osmotic stress at polyethylene glycol (PEG) concentrations 0, 11, 21, and 32.6%). (B) A plot for important variables in each dataset assessed using the Crone-Crosby distance. The codes of the strains in plot (A) refer to the following strains: Bacillus endophyticus PB3, B. australimaris PB17, B. pumilus PB18, B. safensis PB23, Staphylococcus sciuri PB24, B. altitudinis PB37, B. altitudinis PB46, and B. megaterium PB50. Abbreviations used in plot (B) are indoleacetic acid (IAA), gibberellic acid (GA), abscisic acid (ABA), hydrogen cyanide (HCN), Helminthosporium oryzae inhibition percentage (HO_IP), and Pyricularia oryzae inhibition percentage (PO_IP).