Figure 1.
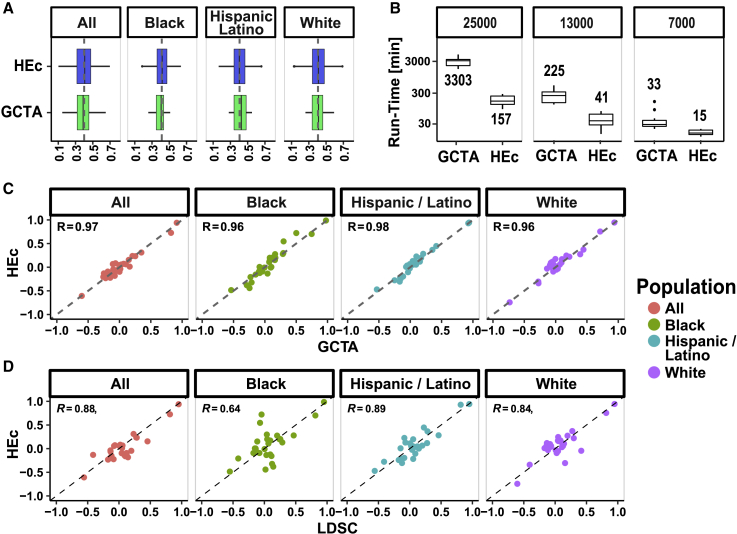
Closed-form HEc is as accurate as GCTA on diverse and admixed populations while being up to 20× faster
Comparison of accuracy and computation speed between our HEc method, GCTA-GREML, and an LD-score-based method (LDSC) based on simulations and the TOPMed dataset. Relatedness data from three self-reported TOPMed backgrounds as well as a combined group of the same size consisting of equal number of all three (n = 7,706) were used to simulate phenotype pairs with known genetic correlation coefficients ().
(A) Boxplots of distributions of estimated from HEc (blue color) and GCTA-GREML (green) when we set = 0.4. See Figure S1 and S2 for multiple simulation values.
(B) Comparison of runtime between HEc and GCTA-GREML for samples comprised of increasing number of individuals (note the log scale).
(C) Comparison of estimates between HEc and GCTA-GREML for all pairs of eight phenotypes selected from the diverse TOPMed cohort either joint or stratified by self-reported background.
(D) Comparison of estimates between HEc and LDSC for all pairs of eight phenotypes from the diverse TOPMed cohort either joint or stratified by self-reported background. See supplemental information and STAR Methods for description of the methods and GWAS used.