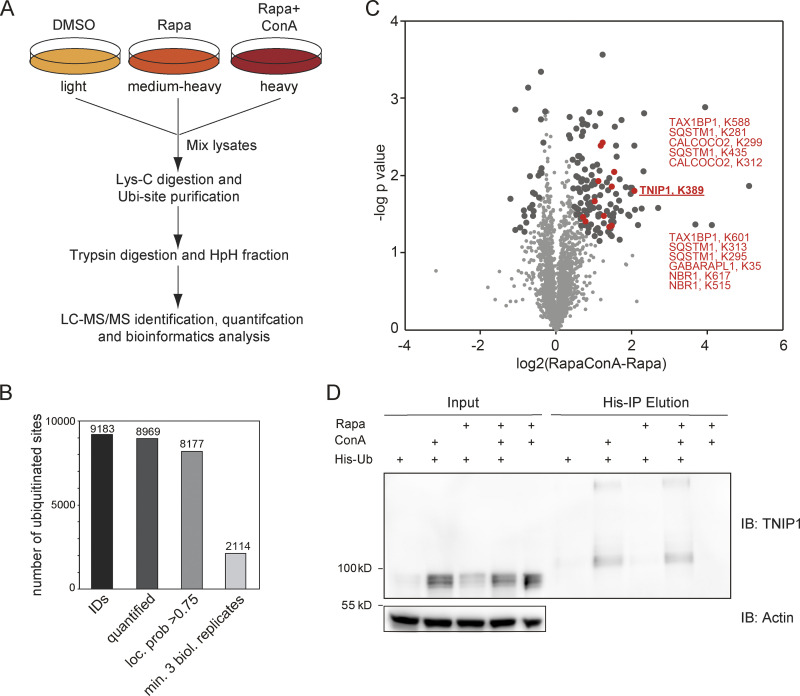
Figure 1.
Ubiquitination and lysosomal degradation of TNIP1. (A) MS workflow to quantify ubiquitination sites potentially involved in autophagy-dependent lysosomal protein degradation. U2OS and HeLa cells were SILAC labeled and treated for 4 h with 100 nM rapamycin (Rapa), Rapa and 2 nM concanamycin A (ConA), or DMSO as control. After mixing of lysates, proteins were digested with Lys-C endoproteinase and ubiquitinated peptides were enriched using the UbiSite approach (Akimov et al., 2018). Enriched peptides were digested with trypsin, followed by high-pH reversed phase fractionation (Batth and Olsen, 2016) and shotgun LC-MS/MS analysis. (B) Detected ubiquitination sites. In three biological replicates, 9,183 ubiquitination sites were identified of which 8,969 were quantified. (C) Volcano plot highlighting significantly regulated ubiquitination sites. Significantly regulated sites comparing Rapa with Rapa + ConA treated cells are highlighted in dark grey (n = 3, paired two-sided t test, FDR < 0.05, S0 = 0.1, 148 sites in total; see Table S1). Data distribution was assumed to be normal but this was not formally tested. Non-regulated sites are colored in light gray. Sites identified on known autophagy receptors are colored in red exemplifying data quality. The newly identified site on TNIP1, K389, is highlighted in bold red. (D) TNIP1 gets ubiquitinated and degraded in the lysosome. U2-OS-StUbEx cells inducibly expressing His-FLAG-tagged ubiquitin at endogenous levels were used to enrich ubiquitinated proteins (Akimov et al., 2014). Under control conditions as well as under 4 h 100 nM rapamycin treatment TNIP1 got ubiquitinated as shown by anti-TNIP1 immunoblots. Ubiquitinated TNIP1 was stabilized by the addition of concanamycin A indicating its lysosomal degradation in treated and nontreated cells. The same was observed for starved cells (HBSS treatment; see Fig. S1). Actin was used as loading control. Note: Due to the design of the experiment and the used gels, the molecular weight marker (PageRuler Plus, #26619; Thermo Fisher Scientific) and the Rapa+/ConA+ sample were run in the same lane. In the respective source data, an additional replicate is shown in which samples ran separately. Source data are available for this figure: SourceData F1.