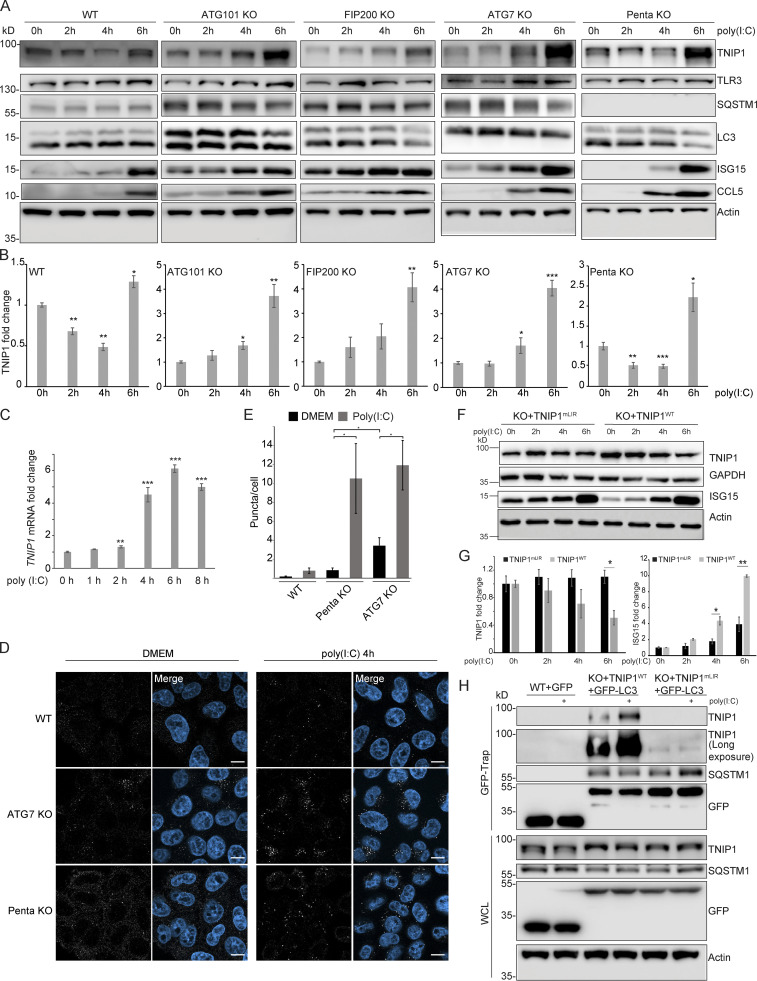
Figure 6.
Poly(I:C) stimulation induces LIR-dependent, specific degradation of TNIP1 by autophagy. (A) Poly(I:C) treatment leads to time-dependent changes in TNIP1 abundance. Poly(I:C) stimulation leads to an autophagy dependent and SLR independent decrease of TNIP1 abundance within the first 4 h as indicated by a block of degradation in ATG101, FIP200 and ATG7 KO cells. Autophagy receptors appear to have a minor influence as degradation still occurs in pentaKO cells. (B) Quantification of blots shown in A (n = 3). Error bars indicate SEM. * = P < 0.05, ** = P < 0.01, *** = P < 0.001 unpaired, two-sided t test compared to 0 h values of respective cell lines. (C) After 2–4 h of poly(I:C) treatment TNIP1 transcription is significantly upregulated. Bar diagrams show quantification of three biological replicates (n = 3), error bars: SEM. * = P < 0.05, ** = P < 0.01, *** = P < 0.001; unpaired, two-sided t test. (D) Representative immunofluorescent images showing endogenous TNIP1 response to poly(I:C) in WT, ATG7 KO, and pentaKO. Cells were either left untreated or treated with 5 µg/ml poly(I:C) for 4 h. Scale bar = 10 µm. (E) Quantification of images shown in D, error bars indicate SEM. * = P < 0.05, unpaired two-sided t test. (F and G) The degradation of TNIP1 depends on functional LIR motifs. TNIP1 WT is degraded in a time-dependent fashion after poly(I:C) stimulation. The double LIR mutant TNIP1 (LIR1+2, TNIP1mLIR) is spared from degradation. Note: Protein amounts of TNIP1 and ISG15 correlate inversely. Due to ectopic expression of TNIP1 variants regulation based on transcriptional/translational control as shown in A is lost. E shows quantification of blots exemplified in D (n = 3). Error bars indicate SEM. * = P < 0.05, unpaired, two-sided t test. KO1 cells were used for reconstitution. (H) Poly(I:C) induces a LIR-dependent interaction with LC3. Indicated HeLa cells expressing GFP-LC3 were used for anti-GFP AP. Cells expressing TNIP1mLIR do not exhibit an increased interaction between TNIP1 and GFP-LC3 after poly(I:C) treatment, in contrast to cells expressing TNIP1WT. KO1 cells were used for reconstitution. In B, C, E, and G, data distribution was assumed to be normal, but this was not formally tested. Source data are available for this figure: SourceData F6.