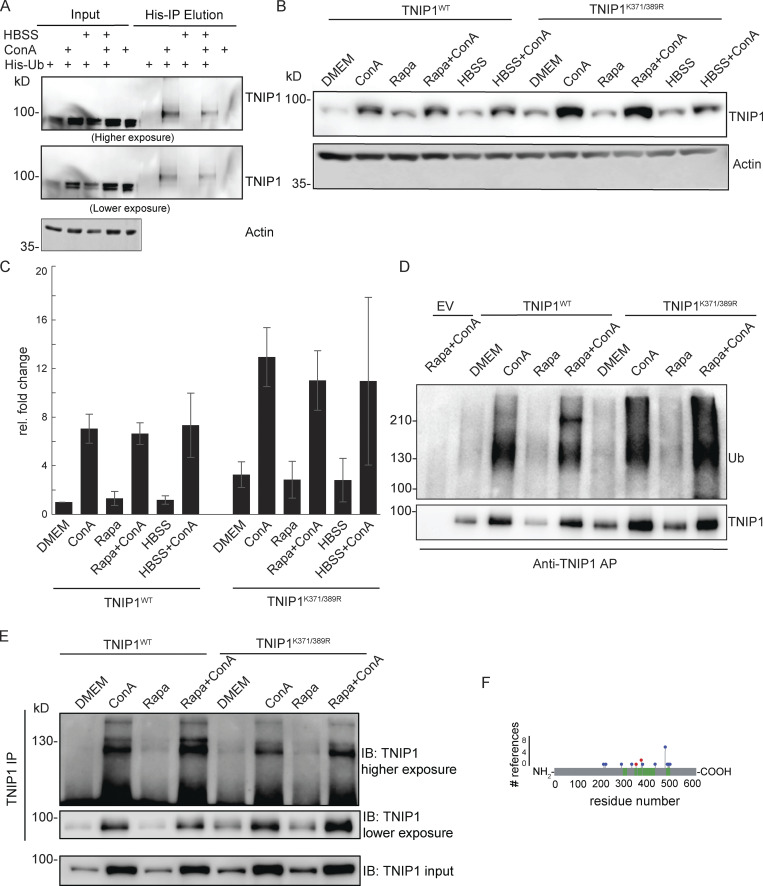
Figure S1.
TNIP1 gets ubiquitinated and degraded in the lysosome. (A) U2-OS-StUbEx cells inducibly expressing His-FLAG-tagged ubiquitin at endogenous levels were used to enrich ubiquitinated proteins (Akimov et al., 2014). Under control conditions as well as under starvation treatment (HBSS), TNIP1 gets ubiquitinated as shown by anti-TNIP1 immunoblots. Ubiquitinated TNIP1 was stabilized by the addition of concanamycin A (ConA) indicating its lysosomal degradation in treated and nontreated cells. Actin was used as loading control. (B and C) Mutations of identified TNIP1 ubiquitination sites do not lead to reduced lysosomal degradation as indicated by stabilized protein amounts by ConA treatment. This is the case for fed control conditions (DMEM) as well as under active autophagy (Rapa and HBSS treatment). C shows quantification of blots exemplified in B (n = 3, error bars indicate SD). (D and E) Mutated TNIP1K371/389R is still getting ubiquitinated as indicated by anti-TNIP1 IP followed by anti-ubiquitin (D) and anti-TNIP1 (E) Western blot. The addition of ConA leads in all cases to a stabilization of non-ubiquitinated and polyubiquitinated protein variants. (F) Identified ubiquitination sites according to PhosphoSitePlus database and this study. Gray bar depicts the amino acid sequence of TNIP1. Sections in green mark tryptic peptides identified in this study, i.e., sequence coverage of TNIP1. Amino acids marked in blue highlight published ubiquitination sites, number of references shown on y-axis. Amino acids marked in red were identified in this study. Source data are available for this figure: SourceData FS1.