Abstract
Objective
Besides breast and gastric cancer, HER2 amplification/mutation are also found in lung adenocarcinoma (LUAD). However, the correlation between HER2 variations and the phenotype of immunogenicity and tumor immune microenvironment (TIME) in LUAD compared with breast and gastric cancer has yet to be fully elucidated.
Methods
We integrated public databases (discovery set) and internal data (validated set) of 288 patients representing three distinct HER2-altered tumors. Genomic data were used to identify somatic mutations, copy number variations, and calculate tumor mutational burden (TMB) and microsatellite instability score. RNA sequencing was conducted to estimate immune gene signatures and contents of tumor-infiltrating immune cell populations. Finally, IHC was used to determine PD-L1 expression and the tumoral-infiltration of immune cells in 50 HER2-variant tumor specimens with no prior therapeutic regimens.
Results
Compared with HER2-amplified breast and gastric cancers, patients with HER2-amplified LUAD showed higher immunogenicity, mainly manifested in immune checkpoints expression and tissue/blood TMB. Additionally, HER2-amplified LUAD exhibited an inflamed TIME with remarkably increased genes encoding HLAs, T-cell activity and immune cell-type, and accompanied with tumor‐infiltrating lymphocytes. In LUAD, patients with HER2 amplification possessed higher tissue TMB than HER2 mutation, whereas no difference was observed in PD-L1 expression. HER2 amplification (primary) was associated with significantly higher PD-L1 expression and TMB than acquired HER2 amplification after resistance to EGFR-TKIs.
Conclusion
Patients with HER2-amplified LUAD have better immunogenicity and/or an inflamed TIME among HER2-aberrant tumors. Our study may provide clues for establishing the benefits and uses of ICIs for patients with this disease.
Keywords: HER2, amplification, lung adenocarcinoma, immunogenicity, tumor immune microenvironment
Introduction
Human epidermal growth factor receptor 2 (ERBB2, encoding HER2) is an oncogenic driver that acts as an “orphan” due to the lack of any known EGF family ligand, which is poised to heterodimerize with other ERBB family members to mediate cell proliferation through the Ras-Raf-MAPK and PI3K/Akt signaling pathways (1). HER2 amplification and mutations are two distinct gene alterations that are also observed in tumors other than breast and gastric cancers, such as LUAD and colorectal cancer (2). HER2, also a well-established therapeutic target, is amplified/overexpressed in 12%–20% of breast cancers and 7%–25% of gastroesophageal cancers, for which trastuzumab-based anti-HER2 therapy has transformed the standard of care with a demonstrated survival benefit (3). HER2 amplification and mutations are also found in approximately 2-10% of non-small cell lung cancer (NSCLC). HER2 mutation is recognized as an oncogenic driver in LUAD but the role of HER2-amplified is doubtful (4, 5). HER2 has been shown to participate in the pathophysiology of LUAD, implicating its role as an actionable driver in lung cancers and correlating with poor prognosis (6). In addition, acquired amplification of HER2 has been proposed as a mechanism of resistance to EGFR/ALK-TKIs, which further confirms its role in tumorigenesis (7). In clinical trials to date, neither trastuzumab nor EGFR/HER2-TKIs have produced clinical benefits in HER2-positive NSCLC. Although several novel compounds, such as trastuzumab deruxtecan (T-DXd, DS-8201) and poziotinib, have emerged in recent years, increasing the objective response rate (ORR) to approximately 50% for patients with HER2-mutant NSCLC (8, 9), there is currently no approved targeted therapy for HER2-amplified LUAD. In view of lacking effective treatment options, there is an urgent need to explore new treatment strategies.
Immune checkpoint inhibitors (ICIs) reinvigorate antitumor immune responses by targeting PD-1/PD-L1 pathways and show remarkable clinical efficacy in driver oncogene-negative NSCLC patients. However, it is generally believed that the response is considerably less frequent in oncogene-addicted NSCLC patients, particularly in patients harboring EGFR/ALK variations (10–12). In the first line or above, ICIs among EGFR-mutated NSCLC patients showed almost no response and outcomes were far inferior to those of negative driver oncogenes patients (median overall survival 9.8 vs. 16.3 months). However, heterogeneity in response to immunotherapy may exist across different oncogenes in NSCLC. Data from a limited number of patients have shown that ICIs are feasible as monotherapy or combination among HER2-mutant lung cancer, with an ORR of 16%-52% and a median progression-free survival (PFS) of 4-6 months (13). However, the place taken ICIs against advanced NSCLC harboring HER2 amplification remains undetermined. Available evidence shows controversial results in the introduction of ICIs in different HER-2 amplified tumors. ICIs have generated robust clinical benefits in advanced HER2-amplified gastric cancer, whereas there is no significant benefit in HER2-amplified breast cancer (14, 15). A higher degree of heterogeneity among tumor types and genomic alteration status obscures our insight into whether HER2-aberrant NSCLC and which HER2 genomic variations would benefit from immunotherapy.
Accumulating evidence suggests that PD-L1 expression, TMB, mismatch repair deficient (dMMR)/microsatellite instability (MSI), immune-related gene expression profiles (GEPs), and tumor‐infiltrating lymphocytes (TILs) represent immunogenicity and TIME features, and correlate with the response to ICIs (16–18). Here, to identify whether immunotherapy has a role in HER2-amplified LUAD, we explored and evaluated immunogenicity and TIME traits among breast, lung and gastric tumors with HER2 amplification. We conducted an integrative analysis that incorporated PD-L1 expression, TMB, MSI status, immune-related and immune cell-type GEPs and measures of TILs from cohorts of TCGA database as well as internal data and patient tumor specimens from 3 hospitals.
Materials and methods
Patients selection
Publicly available simple nucleotide variation (SNP), copy number variation (CNV) and transcriptome profiling data from 3 different cohorts were directly downloaded from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/repository) dataset: breast invasive carcinoma (BRCA, n =1111), lung adenocarcinoma (LUAD, n = 554), stomach adenocarcinoma (STAD, n = 442).
As a validation set, we collected 298 patients from more than 60 medical institutions in China whose tumor specimens and/or peripheral blood underwent next-generation sequencing (NGS) from February 2019 to February 2021. Comprehensive genomic profiling was screened according to the following criteria: a) known or confirmed pathologic diagnosis of LUAD, STAD and BRCA. b) identified as tier I variants with strong clinical significance. c) exclusion of germline mutations by pairing peripheral blood samples. Ultimately, 288 patients with LUAD, STAD and BRCA were enrolled in our cohort. Additionally, 61% (176/288) of patients’ tumor tissue was tested for PD-L1 expression by Dako 22C3/Ventana SP142. Detailed information including demographic and clinical information is provided in Table 1 .
Table 1.
Characteristics of patients in the validation set.
| Characteristics | BRCA n=58 | STAD n=55 | LUAD1 n=58 | LUAD2 n=54 | LUAD3 n=63 |
|---|---|---|---|---|---|
| Age, median (range) | 52 (26-79) | 61 (32-80) | 61 (45-91) | 58 (29-87) | 61 (32-85) |
| Gender | |||||
| Female | 58 (100) | 13 (24) | 18 (31) | 25 (46) | 40 (63) |
| Male | 0 (0) | 42 (76) | 40 (69) | 29 (54) | 23 (37) |
| Smoking history | |||||
| Current or former | 8 (14) | 29 (53) | 35 (60) | 17 (31) | 15 (24) |
| Never | 26 (45) | 20 (36) | 13 (23) | 21 (39) | 35 (55) |
| Unknown | 18 (31) | 6 (11) | 10 (17) | 16 (30) | 13 (21) |
| Treatment history | |||||
| Chemotherapy | 9 (15) | 10 (18) | 3 (5) | 0 (0) | 0 (0) |
| Targeted therapy | 1 (2) | 0 (0) | 0 (0) | 5 (9) | 56 (89) |
| Chemo+ Target | 19 (33) | 8 (14) | 0 (0) | 1 (2) | 7 (11) |
| Immunotherapy | 0 (0) | 1 (2) | 0 (0) | 1 (2) | 0 (0) |
| Not-received | 21 (36) | 29 (53) | 45 (78) | 36 (67) | 0 (0) |
| Unknown | 8 (14) | 7 (13) | 10 (17) | 11 (20) | 0 (0) |
| Specimen Type | |||||
| Tissue Blood |
56 (97) 2 (3) |
46 (84) 9 (16) |
50 (86) 8 (14) |
51 (94) 3 (6) |
40 (63) 23 (37) |
| Sequencing Platform | |||||
| HiSeq X Ten | 40 (69) | 47 (85) | 35 (60) | 23 (42)4 | 44 (70) |
| NovaSeq6000 | 18 (31) | 8 (15) | 23 (40) | 19 (35) | 19 (30) |
| HER2 status | |||||
| CNA | 58 (100) | 55 (100) | 58 (100) | – | 63 (100) |
| 20 exon insertion | – | – | – | 42 (87)5 | – |
| Co-EGFR mut | – | – | – | – | 62 (98)6 |
| PD-L1 IHC | 51 (88) | 35 (64) | 35 (60) | 28 (52) | 27 (43) |
| TMB detection | 51 (88) | 45 (82) | 33 (57) | 30 (56) | 24 (38) |
| MSI detection | 51 (88) | 45 (82) | 38 (67) | 28 (52) | 27 (43) |
| MSI-H | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| HLA-locus7 | 51 | 51 | 32 | 13 | 23 |
| Heterozygosity | 38 (75) | 39 (76) | 26 (81) | 6 (46) | 18 (78) |
| Homozygous | 13 (25) | 12 (24) | 6 (19) | 7 (54) | 5 (22) |
All values are n (%), unless otherwise specified.
BRCA, breast invasive carcinoma; STAD, stomach adenocarcinoma; LUAD, lung adenocarcinoma; CNA, copy number amplification; mut, mutation.
1 HER2 amplification.
2 HER2 mutation.
3 HER2-acquired amplification.
412 patients detected HER2 mutation status by ADx-Amplification Refractory Mutation System (ADx-ARMS).
512 patients harboring HER2 p.S310F.
662 patients received anti-EGFR TKIs; 1 patient received anti-ALK TKIs.
7HLA I-heterozygosity, all of the three HLA-I loci (A, B, or C) were heterozygous; HLA I-homozygosity, at least one HLA-I locus (A, B, or C) was homozygous. Patients with HLA I-heterozygous were associated with better survival than HLA I-homozygous when receiving immunotherapy.
To assess PD-L1 expression and immune cell infiltration, we obtained 50 eligible patients’ paraffin blocks from three medical institutions (The First Affiliated Hospital of Xi’an Jiaotong University, Tangdu Hospital and Shaanxi Provincial People’s Hospital), according to the following criteria: a) primary tumors, excluding metastatic sites, b) not received chemotherapy/radiotherapy or other prior to diagnoses, and c) all paraffin blocks were available from January 2020 to May 2021. The clinicopathologic characteristics of the patients are shown in Table 2 .
Table 2.
Characteristics of patients enrolled for IHC analysis.
| BRCA | STAD | LUAD1 | LUAD2 | |
|---|---|---|---|---|
| Characteristics | n=10 | n=10 | n=12 | n=18 |
| Age, median (range) | 58 (44-70) | 63 (39-78) | 57 (44-76) | 53 (31-71) |
| Sex | ||||
| Female | 10 (100) | 5 (50) | 2 (17) | 5 (28) |
| Male | 0 (0) | 5 (50) | 8 (83) | 13 (72) |
| Smoking history | ||||
| Current or former | 1 (10) | 5 (50) | 7 (58) | 8 (44) |
| Never | 7 (70) | 2 (20) | 3 (25) | 8 (44) |
| Unknown | 2 (20) | 3 (30) | 2 (17) | 2 (11) |
| ECOG performance status | ||||
| 0-1 | 10 (100) | 8 (80) | 9 (75) | 14 (78) |
| 2-4 | 0 (0) | 2 (20) | 3 (25) | 4 (22) |
| Stage (TNM) | ||||
| I | 4 (40) | 0 (0) | 1 (8) | 4 (22) |
| II | 3 (30) | 1 (10) | 2 (17) | 5 (28) |
| III | 2 (20) | 5 (50) | 3 (25) | 2 (11) |
| IV | 1 (10) | 4 (40) | 6 (50) | 7 (39) |
| Adjuvant therapy | ||||
| None | 1 (10) | 0 (0) | 1 (8) | 5 (28) |
| Chemotherapy | 0 (0) | 6 (60) | 3 (25) | 6 (33) |
| Chemo + Target | 8 (80) | 0 (40) | 0 (0) | 0 (0) |
| First line therapy | ||||
| Immunotherapy | 0 (0) | 1 (10) | 1 (8) | 1 (6) |
| Chemo + Target | 1 (10) | 2 (20) | 4 (33) | 4 (22) |
| Others | 0 (0) | 1 (10) | 3 (25) | 2 (11) |
| HER2 (IHC) | ||||
| 0 | 0 (0) | 0 (0) | 0 (0) | 5 (28) |
| 1+ | 0 (0) | 0 (0) | 3 (25) | 7 (39) |
| 2+ | 4 (40) | 4 (40) | 4 (33) | 4 (22) |
| 3+ | 6 (60) | 6 (60) | 5 (42) | 2 (11) |
| HER2 status | ||||
| FISH ratio≥ 2 | 10 (100) | 10 (100) | – | – |
| CN≥ 3 | – | – | 12 (100) | 2 (11) |
| 20 exon insertions | – | – | 0 (0) | 18 (100) |
All values are n (%) unless otherwise specified.
BRCA, breast invasive carcinoma; STAD, stomach adenocarcinoma; LUAD, lung adenocarcinoma; ECOG, Eastern Cooperative Oncology Group; Chemo, chemotherapy.
1 HER2 amplification.
2 HER2 mutation.
Sequencing data analysis
For the TCGA cohort, the masked somatic mutation data were analyzed and visualized using the R package maftools (19). GISTIC 2.0 was used to analyze the downloaded CNV segments. As previously described in detail, A threshold of > 0.2 or < −0.2 is a filtering criterion in the segment mean value for amplification or deletion, respectively (20, 21).
For the validation set, tissue genomic DNA paired with peripheral blood genomic DNA followed the process below: 1) sheared into 150-200 bp fragments; 2) constructed DNA and cell-free DNA libraries; 3) sequenced on an Illumina platform; 4) mapped to the human genome assembly: GRCh37/hg19; 5) filtered with public online genome databases: Exome Aggregation Consortium (ExAC), Genome Aggregation Database (gnomAD) and 1000 Genomes Project (1000G); and 6) identified somatic genome variations. We defined somatic mutation frequency > 1% and gene copy number > 3 as clinically significant (21–23). Additionally, OptiType was used to determine the HLA-I loci (24, 25).
TMB and MSI analysis
TMB was defined as the total number of somatic nonsynonymous per megabase of tumor tissue which included gene coding errors, base substitution insertions or deletions in detected coding regions. As previously described in detail (26), in the TCGA cohort, TMB was equal to the raw mutation count divided by 38 Mb (the estimate of the exome size). In the validation set, TMB was equal to the number of single-nucleotide variants (somatic nonsynonymous with depth > 100X and allele frequency ≥ 0.05) detected on 1 Mb of the genome (23). As previously described in detail (27), a total of 134 microsatellite loci with a 15-30 bp span were used to calculate the MSI score. Quality control required over 40 loci to pass. The MSI score is equal to the number of unstable loci divided by the number of loci passing quality control, if ≥ 0.3, MSI-H; if < 0.3, MSS/MSI-L.
Gene expression profiling analyses
In the TCGA cohort, following previous experimental procedures, RNA-sequencing (RNA-seq) data were analyzed using the R package edgeR and normalized by log2-transformation (28). The mRNA expression levels of immune gene signatures, including immune checkpoints, MHC-class-I/II signatures, T cell–inflamed gene expression profiles (GEPs) and immune cell GEPs, were compared among the TCGA-LUAD, TCGA-STAD, and TCGA-BRCA cohorts using the R package limma and were quantified as log2 fragments per kilobase million (FPKM).
The CIBERSORT algorithm, a deconvolution tool with default parameters (29), used a knowledgebase of GEPs and linear support vector regression to estimate the contents of the immune cell expression matrix among the LUAD, STAD and BRCA cohorts.
IHC
Immunostaining was performed on a Ventana BenchMark XT automated IHC stainer (Roche, Basel, Switzerland) according to the manufacturers’ protocols. The primary antibodies were HER2 (clone 4B5, Roche, Basel, Switzerland); PD-L1 (clone SP263, Roche, Basel, Switzerland); CD4 (clone 2H4A2, Proteintech, Wuhan, China); CD8 (clone C8/144B, Cell Signaling Technology, Danvers, MA, United States); FOXP3 (clone 236A/E7, Abcam, Cambridge, United Kingdom); and CD68 (clone KP1, Maxim-Bio, Fuzhou, China), visualized by the OptiView DAB IHC detection kit (Roche). The stained slides were evaluated separately by two specialized oncologic pathologists blinded to the clinical parameters.
Except for nuclear staining of Foxp3, all markers were positive for membranous staining. The expression of PD-L1 and HER2 was manually assessed according to clinical diagnosis criteria (30, 31). For immune cell analysis, all section images were scanned using a Leica SCN400 slide scanner (Leica Microsystems). The intratumoral regions were evaluated for the density of immune cells (number/mm2) under a 20× objective lens field (equal to 0.195 mm2) or a 40× objective lens field (equal to 0.0495 mm2). Combined with cell morphology and staining intensity, the number of positive immune cells was counted at three to five hot spots and calculated as the average (32, 33).
Statistical analysis
Data analysis was conducted using R software (version 4.04) and GraphPad Prism (version 9.0.0). Parametric (Student’s t-test and one-way ANOVA) or nonparametric (Mann-Whitney U test and Kruskal-Wallis test) tests were applied depending on whether the data followed Gaussian distribution. Categorical variables were analyzed by the Chi-square test or Fisher exact test. A two-tailed P <0.05 was considered statistically significant unless otherwise specified. Additionally, the ggplot2 R package was applied to visualize the boxplot and violinplot, the ComplexHeatmap R package was used to visualize the Heatmap and the fmsb R package was used to visualize the Radar Chart.
Results
Correlation between HER2 amplification and PD-L1/other immune checkpoints expression in LUAD patients compare to BRCA and STAD
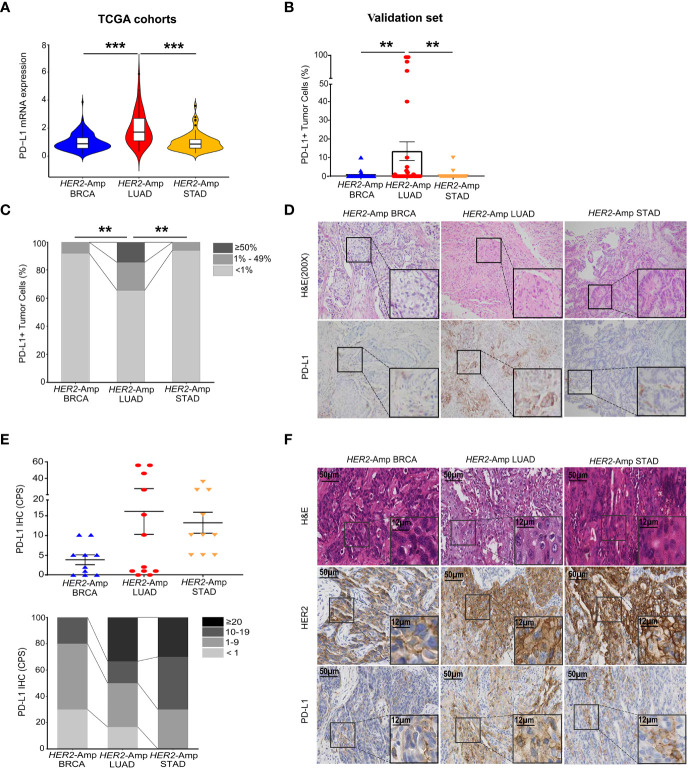
PD-L1 expression has high predictive value in guiding cancer immunotherapy (17). Thus, we first investigated PD-L1 expression levels between HER2 amplification and PD-L1 expression in BRCA, LUAD and STAD cohorts by interrogating and analyzing RNA-seq data from TCGA. The level of PD-L1 mRNA expression in HER2-amplified LUAD was significantly higher than that in HER2-amplified STAD and BRCA cohorts ( Figure 1A ). However, no differences were noted between HER2-amplified STAD and BRCA cohorts. Further analysis confirmed this result at the PD-L1 protein level ( Figures 1B–D ).
Figure 1.
Correlation between HER2 amplification and PD-L1 and/or other immune checkpoint expressions in patients with BRCA, LUAD and STAD. (A) PD-L1 mRNA expression among the HER2-amplified BRCA cohort (n = 212), HER2-amplified LUAD cohort (n = 56) and HER2-amplified STAD cohort (n = 71) based on analysis of the TCGA database. (B, C) Statistical results of PD-L1 staining in HER2-amplified cohorts of 51 BRCA patients, 35 LUAD patients and 35 STAD patients. PD-L1 expression in tumor cells is classified as ≥ 50%, 1% to 49% and < 1%. (D) Representative images of hematoxylin-eosin (HE) staining and PD-L1 IHC staining in BRCA, LUAD and STAD tissues with HER2 amplification. (E, F) Statistical analysis (E) and images (F) of HE, HER2 and PD-L1 IHC staining were conducted. PD-L1 expression was assessed at cut-offs of ≥ 20, 10 to 19, 1 to 9 and < 1 based on the combined positive score (CPS). CPS = [(number of PD-L1-positive tumor cells and mononuclear inflammatory cells)/(total number of tumor cells)] × 100. Amp, amplified; BRCA, breast invasive carcinoma; LUAD, lung adenocarcinoma; STAD, stomach adenocarcinoma. P < 0.05 was regarded as significantly different. ***P < 0.001, **P < 0.01.
Except for tumor cells, PD-L1 is also expressed in immune cells. To eliminate the singleness of the PD-L1 evaluation method and further confirm the association between HER2 amplification and PD-L1 expression as the TCGA cohorts and validation set demonstrated, we detected 10 BRCA, 12 LUAD, 10 STAD surgical or biopsy specimens using IHC. Immunostaining showed that HER2-amplified LUAD specimens tended to have stronger staining for the PD-L1 protein assessed by the combined positive score (CPS, Figure 1E ). In addition, STAD harboring HER2 amplification equally expressed higher PD-L1 owing to its immune cell staining within the tumor stroma. However, HER2-amplified BRCA continued to lower the expression of PD-L1 when assessing both tumor and immune cells ( Figure 1F ).
To figure out the association between HER2 amplification and other non–PD-L1 immune checkpoints in BRCA, LUAD and STAD patients, we next exploit RNA-seq data to depict the expression levels of 7 key immune checkpoints. A heatmap displayed remarkably increased expression of checkpoints in the HER2-amplified LUAD cohorts while decreased expression in the HER2-amplified BRCA and STAD cohorts. More interestingly, PD-L1 mRNA expression was markedly increased in the HER2-amplified LUAD subgroup, relative to other immune inhibitory checkpoints. Finally, boxplots were constructed to represent the other two most significantly different immune checkpoints (PD-1 and IDO1; Figures S1A, S1B ).
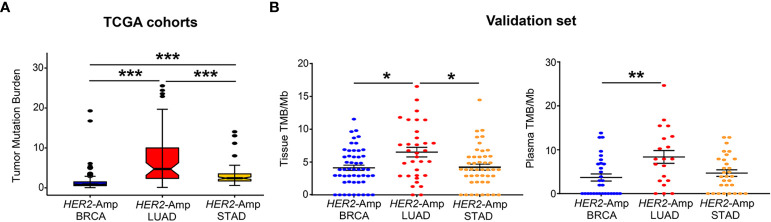
Correlation between HER2 amplification and TMB in LUAD patients compare to BRCA and STAD
TMB is an independent predictor/indicator of response to immunotherapy in pan-cancers (34). From TCGA cohort analysis, patients with HER2-amplified LUAD showed significantly higher tissue TMB than HER2-amplified BRCA and STAD cohorts ( Figure 2A ). Consistent with the finding from the discovery set, our validation set manifested a similar result: HER2- amplified LUAD patients had higher tissue TMB and/or blood TMB than the other two cohorts, particularly HER2-amplified BRCA cohorts ( Figure 2B ).
Figure 2.
Correlation between HER2 amplification and TMB in patients with BRCA, LUAD and STAD. (A) Tissue TMB driven by HER2 amplification among three tumors from TCGA cohorts. (B) Tissue and blood TMB driven by HER2 amplification among three tumors from the validation set. TMB, tumor mutation burden, was defined as the total number of somatic nonsynonymous per megabyte. P < 0.05 was regarded as significantly different. ***P < 0.001, **P < 0.01, *P < 0.05.
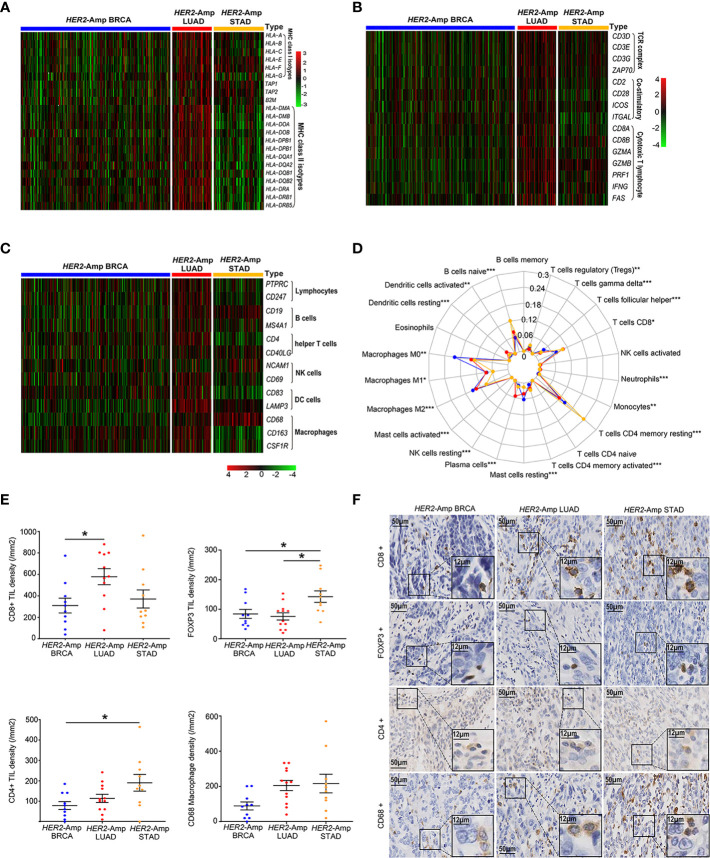
The landscape of the TIME among HER2-amplified BRAC, LUAD and STAD
Anti-tumor immunity requires antigen presentation, and T cell priming and trafficking to the tumor tissue. These steps require the coordinated activity of immune networks within the TIME. We used TCGA-RNAseq data and applied the IHC method to depict the whole process as comprehensively as possible. Compared with the other two cohorts, genes encoding classical MHC class I/II antigens and related antigen processing machinery proteins (TAP1 and B2M) were expressed at significantly higher levels in HER2-amplified LUAD cohorts ( Figure 3A ). Additionally, HER2-amplified LUAD possessed increased immune-related GEPs ( Figure 3B ), such as T-cell antigen receptor (CD3), costimulatory molecules (CD28, ICOS, etc.) and cytotoxic effect-related genes (CD8A, IFNG, GZMA, etc.). To sufficiently assess the contents of immune cell infiltration, CIBERSORT algorithm and immune cell-related genes were used to quantify various immune populations ( Figures 3C, D ). The absolute number of multiple immune cell populations, including CD4+, CD8+ T cells and M1 macrophages, were markedly increased in HER2-amplified LUAD. Additionally, regulatory T cells (Tregs), a type of suppressive T cell, were lower infiltration in HER2-amplified LUAD cohorts.
Figure 3.
The landscape of the TIME among HER2-amplified BRAC, LUAD and STAD. (A) Heatmap depicting the expression of HLA-related gene profiles among three distinct tumors from RNA-seq. (B) Heatmap depicting the process of T cell activation, including antigen recognition, signal transduction and T cell immune efficacy from RNA-seq data. (C) Heatmap depicting the mRNA expression levels of the immune cell-associated gene signature. (D) A radar chart displaying the fraction of infiltrated immune cells by CIBERSORT. The blue, red and yellow lines represent HER2-amplified BRAC, LUAD and STAD, respectively. (E) Comparison of TIL and macrophage densities at intratumoral regions among patients with HER2-amplified LUAD (n=12), HER2-amplified BRCA (n=10) and HER2-amplified STAD (n=10). Note: The paraffin section of 1 patient with HER2-amplified LUAD failed the quality control after CD8 IHC staining and was not included in the statistical analysis. (F) Representative images of HE staining and IHC staining of HER2, CD4, CD8, FOXP3 and CD68 among the above-mentioned tumors. P < 0.05 was regarded as significantly different. ***P < 0.001, **P < 0.01, *P < 0.05.
To further make our preliminary results more convincing, we conducted IHC to evaluate the density of immune cell infiltration among patients with HER2-amplified BRCA, LUAD and STAD. As expected, we observed increased CD8+ TIL densities and decreased FOXP3+ TIL densities in HER2 amplified LUAD specimens compared to HER2-amplified BRCA and STAD specimens. In addition, we found increased CD4+ TILs densities in HER2-amplified STAD specimens, corresponding to upregulation of FOXP3+ Tregs. The densities of CD68 macrophages were similar between the HER2-amplified STAD and LUAD groups ( Figures 3E, F ). Regardless of the subtype of immune cells, HER2-amplified BRCA exhibited lower TIL and macrophage densities than the other two groups. Generally, the IHC results are consistent with the RNA-seq analysis.
The immunogenicity and TME in HER2-aberrant LUAD
Next, we investigated the PD-L1, TMB and immune cell infiltrations between HER2 amplification and mutation in LUAD. A significantly higher TMB was apparent for patients with HER2 amplification, but no difference was observed in PD-L1 expression ( Figures 4A–D ). We next evaluated immune cell densities in patients with HER2 amplification and HER2 mutation. Although no significant difference was obtained regarding the TIL and macrophage densities, there was a tendency for both CD8+ and CD4+ TIL counts to be higher in tumors harboring HER2 amplification than in those harboring HER2 mutation ( Figures 4E, F ).
Figure 4.
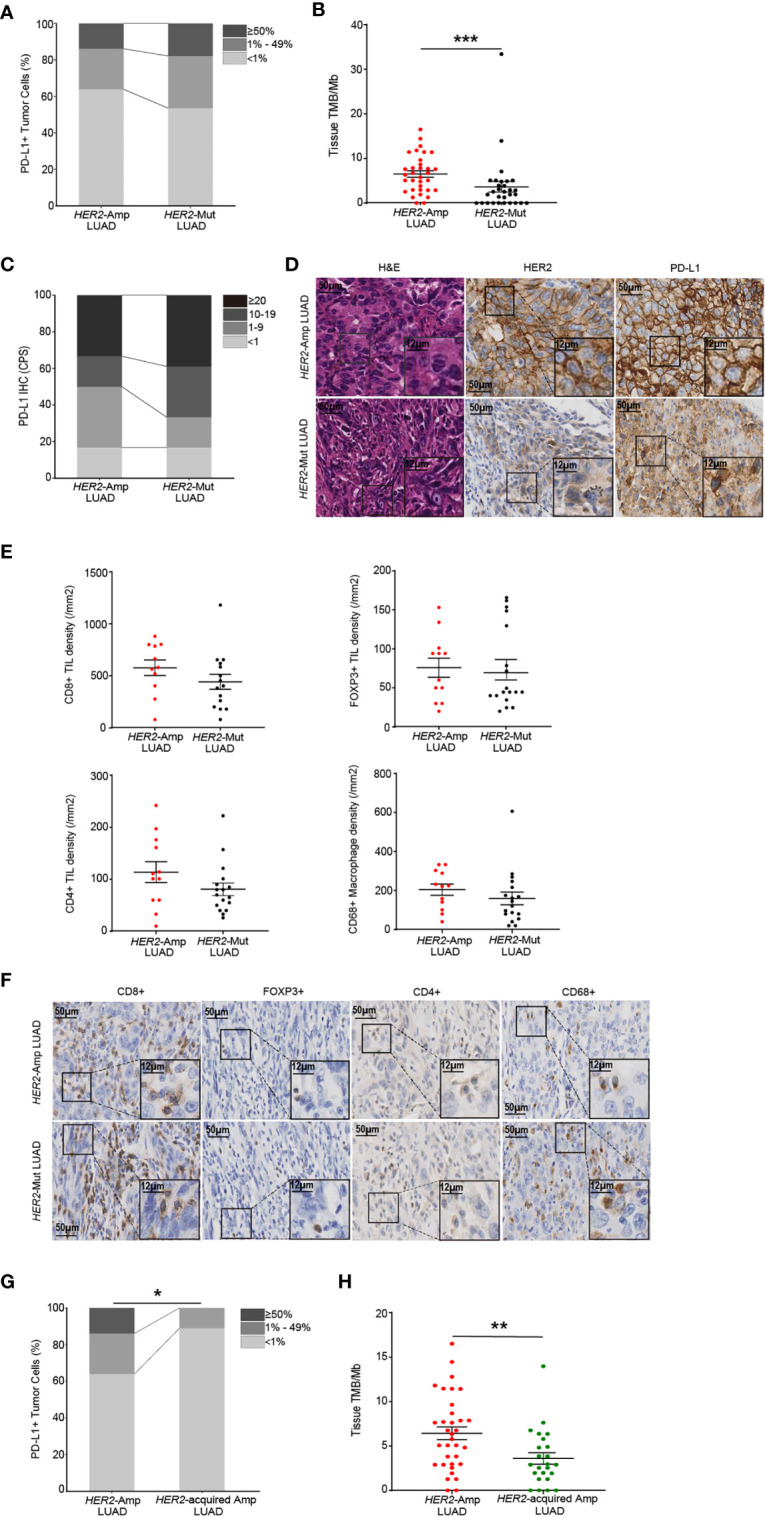
The immunogenicity and TIME in HER2-aberrant LUAD. (A, B) Difference in PD-L1 expression on tumor cells (A) and the level of TMB (B) between amplification and mutation. (C, D) Statistical results (c) and images (D) of PD-L1 expression evaluated by CPS. (E, F) Densities of CD8+, POXP3+ and CD4+ lymphocytes, and CD68+ macrophages (E) between LUAD patients with HER2 amplification (n=12) and mutation (n=18). Corresponding IHC images are shown in (F). Note: The paraffin section of 3 patients with HER2-mutant LUAD failed the quality control after CD8 IHC staining and were not included in the statistical analysis. 1 patient with HER2-mutant LUAD was not included after CD4 IHC staining because of the same problem. (G, H) The PD-L1 (G) and TMB (H) levels in LUAD patients with HER2 amplification and acquired HER2 amplification. P < 0.05 was regarded as significantly different. ***P < 0.001, **P < 0.01, *P < 0.05.
In addition, amplification of HER2 is regarded as one of the resistance mechanisms in NSCLC patients after receiving anti-EGFR/ALK TKIs [7]. We distinguished acquired HER2 amplification through co-mutation status and treatment history and demonstrated that acquired HER2 amplification LUAD rendered lower PD-L1 expression and TMB than HER2 amplification (naïve or primary amplification, Figures 4G, H ). Together, these findings supported that HER2 amplification represents a high degree of immunity and immunogenicity among HER2-aberrant NSCLC.
Discussion
The characteristics of the immunogenicity and TIME in HER2-amplified LUAD have not yet been illustrated. The lower incidence of HER2 amplification or mutation in LUAD leaves challenges in depicting the landscape of immunogenicity and TIME features when compared with wild-type NSCLC patients. Thus, we explored the correlation between HER2 amplification and characteristics of immunogenicity and TIME in LUAD contrast to gastric cancer that benefited from immunotherapy (15), and breast cancer that did not (17), to establish a theoretical feasibility of ICIs for HER2-amplified LUAD. Here, we found that patients with HER2-amplified LUAD showed higher immunogenicity, mainly manifested in PD-L1 expression at the mRNA and protein level, as well as tissue and blood TMB, in comparison with HER2-amplified breast and gastric cancers. Additionally, HER2-amplified LUAD exhibited an inflamed TIME with remarkably increased genes encoding HLAs, T-cell activation and immune cell-type, and accompanied with TILs. In LUAD, we observed that patients with HER2 amplification possessed elevated TMB than those with HER2 mutation, whereas no difference was observed regarding PD-L1 expression. HER2 amplification (naïve or primary amplification) was associated with significantly increased PD-L1 expression and higher TMB than acquired HER2 amplification after resistance to EGFR-TKIs. The pooled results support that HER2-amplified LUAD captures a higher likelihood of deriving benefits from ICIs.
It is well known that NSCLC as a whole has higher PD-L1 expression and TMB relative to breast and gastric cancers, but the whole character could not accurately define the individuals, as EGFR-mutant NSCLC and triple-negative breast cancer (TNBC), which represented the different immunogenicity and generated an opposite immune response to ICIs (10, 35). It is believed that specific oncogenes represent their own unique immunogenicity and TIME within the tumor. As per available literature in NSCLC, higher PD-L1 expression and TMB are frequently observed in male smokers and in patients harboring KRAS mutation, whereas there is no correlation with EGFR alterations (12, 36). We found that PD-L1 expression at the mRNA and protein level, as well as tissue and blood TMB are dominant in HER2-amplified LUAD compared with breast and gastric cancers harboring HER2 amplification. In essence, it can be seen from the literature that HER2-amplified LUAD was associated with male smokers and invasive features (37). An RTDs study confirmed that HER2 amplified NSCLC tumors were associated with higher TMB (100% of tumors≥10 muts/Mb), although the study only involved 6 patients (38). In comparison, prior studies indicated that increased PD-L1 expression and TMB in breast cancer were enriched in TNBC and lobular carcinomas (39). In gastric cancers, HER2 amplification is inversely correlated with PD-L1 expression (40). Bioinformatics research indicated that HER2 amplification was not classified into PD-L1 amplification and hypermutation subtypes of gastric cancer (41). Such findings could be the reason HER2-amplified LUAD shows increased immunogenicity compared with breast and gastric cancer.
Little information is available regarding the correlation between the TIME and HER2 amplification in NSCLC. Conventional opinion is that breast cancers bearing HER2 amplification result in a non-inflamed TIME, with relatively low infiltration of intratumor TILs (39, 42). In contrast, HER2-amplified gastric cancer was dominant in the gastroesophageal junction (GEJ) and intestinal histology, connected with microbial infection, which generates a chronic inflammation status (41, 43). In patients with HER2-amplified LUAD, exposure to accumulated mutation load can release neoantigens that trigger T cell activation and recruit immune cell infiltration. Hence, as we demonstrated, HER2-amplified LUAD samples showed increased immune-related and immune cell-type gene expression profiles (GEPs) compared with the other two cancer groups, especially the HER2-amplified BRCA group. The presence of a greater number of TILs and fewer Tregs has been observed in HER2-amplified LUAD regardless of RNA-seq and IHC results. Of note, the upregulation of CD4+ TILs is accompanied by FOXP3+ Treg infiltrations in HER2-amplified gastric cancer. In general, in comparison with breast and gastric tumors harboring HER2 amplification, these pooled results implicated the possibility that HER2-amplified LUAD establishes a favorable TIME for developing therapeutic efficacy in cancer immunotherapy.
Among HER2-aberrant NSCLC, HER2 mutation mainly occurs in female no-smokers, performing similar clinicopathological characteristics to EGFR mutation, and acquired HER2 amplification is a mechanism of resistance to EGFR-TKIs (7). We observed that patients with HER2 mutation and acquired HER2 amplification possessed lower TMB than HER2 amplification. Prior studies pointed out that the non-inflamed TIME for patients with NSCLC harboring EGFR mutation is thought to be reflective of and caused by their low TMB (42), which naturally accounts for our observation. However, unlike available literature suggesting that HER2-mutant NSCLC has been shown to have lower PD-L1 expression than wild-type NSCLC (44), we discovered that cases with HER2 mutation exhibited higher PD-L1 expression at the protein level, corresponding to dense TILs. This phenomenon is similar to the conflicting results regarding the relationship between PD-L1 and EGFR mutation (45). In addition, we found that patients with acquired HER2 amplification had lower PD-L1 expression than those with primary amplification. Although a recent study indicated that EGFR-TKI treatment was associated with a significant increase in PD-L1 expression in EGFR-mutant NSCLC, the impact of cytotoxic chemotherapy is not excluded and specific to acquired HER2 amplification remains underdetermined (33). In brief, HER2 amplification (naïve or primary amplification) represents a high degree of immunity and immunogenicity among HER2-aberrant LUAD.
The lower frequency of HER2 amplification or mutation in NSCLC imposes restrictions on the analysis of the immunotherapy effect. Our study was necessarily limited in that the efficacy of ICIs targeting NSCLC patients harboring HER2 alterations is lacking. It is generally believed that tumors bearing an inflamed phenotype are conducive to recognition by the immune system and further improve the clinical benefits of immunotherapy. Existing evidence demonstrated that patients with HER2-amplified gastric cancer could benefit from ICIs treatment rather than HER2-amplified breast cancer, and our findings suggest that HER2-amplified LUAD shows higher immunogenicity and a more “inflamed” TIME than HER2-amplified breast cancer and gastric cancer. Thus, we speculate that there are good prospects for practical applications of ICIs in HER2-amplified LUAD that can benefit from immunotherapy. However, the consistency between immune markers and the efficacy of immunotherapy in HER2-amplified LUAD remains elusive and represents an area of further comprehensive evaluation.
In summary, we conducted an assessment of immune-related biomarkers and elucidated an “inflamed” phenotype of HER2-amplified LUAD among HER2-positive tumors, which would provide a theoretical basis for the practical application of ICIs and accelerate the pace toward immuno-precision direction.
Data availability statement
All data generated or analyzed during this study are included in this publish article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of The First Affiliated Hospital of Xi’an Jiaotong University (No.XJTU1AF2021LSK-3379). The work was conducted in accordance with the Declaration of Helsinki. 50 patients who obtained eligible paraffin blocks have signed written informed consent in accordance with the ethical guidelines, their clinicopathological information was captured from the Electronic Medical Record (EMR).
Author contributions
Conceived and designed the analysis: QW, ZM, WL, YJ and HG. Contributed data or analysis tools: SW, LW, LC, ZY, XF, PJ, YB, LX, SZ. Performed the analyses: XJ, LJ, ML and GZ. Wrote the paper: QW, ZM, WL, YH, YJ and HG. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Genecast Biotech Co. (Beijing, China), Simceredx Co. (Nanjing, China) and Burning Rock Biotech Co. (Guangzhou, China) for providing NGS data and helping with bioinformatic analysis, as well as Tangdu Hospital and Shaanxi Provincial People’s Hospital for providing tumor specimens. We also appreciate the support from doctors and patients.
Funding
This work was supported by the Chinese Society of Clinical Oncology MSD Cancer Research (Y-MSD2020-0247) and Key Clinical Foundation of the First Affiliated Hospital of Xi ‘an Jiaotong University (XJTU1AF-CRF-2019-001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1042072/full#supplementary-material
References
- 1. Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. (2005) 5:341–54. doi: 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- 2. Chmielecki J, Ross JS, Wang K, Frampton GM, Palmer GA, Ali SM, et al. Oncogenic alterations in ERBB2/HER2 represent potential therapeutic targets across tumors from diverse anatomic sites of origin. oncologist. (2015) 20:7–12. doi: 10.1634/theoncologist.2014-0234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cen S, Liu Z, Pan H, Han W. Clinicopathologic features and treatment advances in cancers with HER2 alterations. BBA Rev Cancer. (2021) 1876(2):188605. doi: 10.1016/j.bbcan.2021.188605 [DOI] [PubMed] [Google Scholar]
- 4. Zhao J, Xia Y. Targeting HER2 alterations in non–Small-Cell lung cancer: A comprehensive review. JCO Precis Oncol (2020) 4:411–25. doi: 10.1200/PO.19.00333 [DOI] [PubMed] [Google Scholar]
- 5. Li BT, Ross DS, Aisner DL, Chaft JE, Hsu M, Kako SL, et al. HER2 amplification and HER2 mutation are distinct molecular targets in lung cancers. J Thorac Oncol (2016) 11(3):414–9. doi: 10.1016/j.jtho.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu L, Shao X, Gao W, Bai J, Wang R, Huang P, et al. The role of human epidermal growth factor receptor 2 as a prognostic factor in lung cancer: A meta-analysis of published data. J Thorac Oncol (2010) 5(12):1922–32. doi: 10.1097/JTO.0b013e3181f26266 [DOI] [PubMed] [Google Scholar]
- 7. Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, et al. HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFR T790M mutation. Cancer Discovery (2012) 2:922–33. doi: 10.1158/2159-8290.CD-12-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li BT, Smit EF, Goto Y, Nakagawa K, Udagawa H, Mazières J, et al. Trastuzumab deruxtecan in HER2-mutant non-Small-Cell lung cancer. N Engl J Med (2022) 386(3):241–51. doi: 10.1056/NEJMoa2112431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robichaux JP, Elamin YY, Vijayan RSK, Nilsson MB, Hu L, He J, et al. Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell (2019) 36:444–57.e7. doi: 10.1016/j.ccell.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lisberg A, Cummings A, Goldman JW, Bornazyan K, Reese N, Wang T, et al. A phase II study of pembrolizumab in EGFR-mutant, PD-L1+, tyrosine kinase inhibitor naïve patients with advanced NSCLC. J Thorac Oncol (2018) 13:1138–45. doi: 10.1016/j.jtho.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Addeo A, Passaro A, Malapelle U, Luigi Banna G, Subbiah V, Friedlaender A. Immunotherapy in non-small cell lung cancer harbouring driver mutations. Cancer Treat Rev (2021) 96:102179. doi: 10.1016/j.ctrv.2021.102179 [DOI] [PubMed] [Google Scholar]
- 12. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res (2017) 23:3012–24. doi: 10.1158/1078-0432.CCR-16-2554 [DOI] [PubMed] [Google Scholar]
- 13. Saalfeld FC, Wenzel C, Christopoulos P, Merkelbach-Bruse S, Reissig TM, Laßmann S, et al. Efficacy of immune checkpoint inhibitors alone or in combination with chemotherapy inNSCLC harboring ERBB2 mutations. J Thorac Oncol (2021) 16:1952–8. doi: 10.1016/j.jtho.2021.06.025 [DOI] [PubMed] [Google Scholar]
- 14. Janjigian YY, Maron SB, Chatila WK, Millang B, Chavan SS, Alterman C, et al. First-line pembrolizumab and trastuzumab in HER2-positive oesophageal, gastric, or gastro-oesophageal junction cancer: an open-label, single-arm, phase 2 trial. Lancet Oncol (2020) 21:821–31. doi: 10.1016/S1470-2045(20)30169-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emens LA, Esteva FJ, Beresford M, Saura C, De Laurentiis M, Kim SB, et al. Trastuzumab emtansine plus atezolizumab versus trastuzumab emtansine plus placebo in previously treated, HER2-positive advanced breast cancer (KATE2): a phase 2, multicentre, randomised, double-blind trial. Lancet Oncol (2020) 21:1283–95. doi: 10.1016/S1470-2045(20)30465-4 [DOI] [PubMed] [Google Scholar]
- 16. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med (2015) 372:2509–20. doi: 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol (2021) 18:345–62. doi: 10.1038/s41571-021-00473-5 [DOI] [PubMed] [Google Scholar]
- 18. Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, et al. T-Cell-Inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol (2019) 37:318–27. doi: 10.1200/JCO.2018.78.2276 [DOI] [PubMed] [Google Scholar]
- 19. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res (2018) 28:1747–56. doi: 10.1101/gr.239244.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol (2011) 12:R41. doi: 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Dessel LF, van Riet J, Smits M, Zhu Y, Hamberg P, van der Heijden MS, et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nat Commun (2019) 10:5251. doi: 10.1038/s41467-019-13084-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang M, Fan W, Ye M, Tian C, Zhao L, Wang J, et al. Molecular profiles and tumor mutational burden analysis in Chinese patients with gynecologic cancers. Sci Rep-Uk. (2018) 8:8990. doi: 10.1038/s41598-018-25583-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, American society of clinical oncology, and college of American pathologists. J Mol Diagn. (2017) 19:4–23. doi: 10.1016/j.jmoldx.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science (2018) 359:582–7. doi: 10.1126/science.aao4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics (2014) 30:3310–6. doi: 10.1093/bioinformatics/btu548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med (2017) 9:34. doi: 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song Y, Huang J, Liang D, Hu Y, Mao B, Li Q, et al. DNA Damage repair gene mutations are indicative of a favorable prognosis in colorectal cancer treated with immune checkpoint inhibitors. Front Oncol (2021) 10:549777. doi: 10.3389/fonc.2020.549777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods (2015) 12:453–7. doi: 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ventana Medical Systems . PATHWAY anti-HER-2/neu (4B5) rabbit monoclonal antibody package insert, revision f. Available at: https://productlibrary.ventanamed.com/ventana_portal/OpenOverlaySe-rvlet?launchIndex=1&objectId=790-299114427US.
- 31. Ventana Medical Systems . VENTANA PD-L1 (SP263) assay. Available at: https://www.accessdata.fda.gov/cdrh_docs/pdf16/P160046C.pdf.
- 32. Shen YC, Hsu CL, Jeng YM, Ho MC, Ho CM, Yeh CP, et al. Reliability of a single-region sample to evaluate tumor immune microenvironment in hepatocellular carcinoma. J Hepatol (2020) 72:489–97. doi: 10.1016/j.jhep.2019.09.032 [DOI] [PubMed] [Google Scholar]
- 33. Isomoto K, Haratani K, Hayashi H, Shimizu S, Tomida S, Niwa T, et al. Impact of EGFR-TKI treatment on the tumor immune microenvironment in EGFR mutation-positive non-small cell lung cancer. Clin Cancer Res (2020) 26:2037–46. doi: 10.1158/1078-0432.CCR-19-2027 [DOI] [PubMed] [Google Scholar]
- 34. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med (2017) 377:2500–1. doi: 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumaband nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med (2018) 379:2108–21. doi: 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 36. Takada K, Okamoto T, Shoji F, Shimokawa M, Akamine T, Takamori S, et al. Clinical significance of PD-L1 protein expression in surgically resected primary lung adenocarcinoma. J Thorac Oncol (2016) 11:1879–90. doi: 10.1016/j.jtho.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 37. Kim EK, Kim KA, Lee CY, Shim HS. The frequency and clinical impact of HER2 alterations in lung adenocarcinoma. PloS One (2017) 12:e0171280. doi: 10.1371/journal.pone.0171280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dudnik E, Bshara E, Grubstein A, Fridel L, Shochat T, Roisman LC, et al. Rare targetable drivers (RTDs) in non-small cell lung cancer (NSCLC): Outcomes with immune checkpoint inhibitors (ICPi). Lung Cancer. (2018) 124:117–24. doi: 10.1016/j.lungcan.2018.07.044 [DOI] [PubMed] [Google Scholar]
- 39. Ali HR, Glont SE, Blows FM, Provenzano E, Dawson SJ, Liu B, et al. PD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytes. Ann Oncol (2015) 26:1488–93. doi: 10.1093/annonc/mdv192 [DOI] [PubMed] [Google Scholar]
- 40. Angell HK, Lee J, Kim KM, Kim K, Kim ST, Park SH, et al. PD-L1 and immune infiltrates are differentially expressed in distinct subgroups of gastric cancer. Oncoimmunology (2019) 8:e1544442. doi: 10.1080/2162402X.2018.1544442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature (2014) 513(7517):202–9. doi: 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumagai S, Koyama S, Nishikawa H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat Rev Cancer. (2021) 21:181–97. doi: 10.1038/s41568-020-00322-0 [DOI] [PubMed] [Google Scholar]
- 43. Shim JH, Yoon JH, Choi SS, Ashktorab H, Smoot DT, Song KY, et al. The effect of helicobacter pylori CagA on the HER-2 copy number and expression in gastric cancer. Gene (2014) 546(2):288–96. doi: 10.1016/j.gene.2014.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tian P, Zeng H, Ji L, Ding Z, Ren L, Gao W, et al. Lung adenocarcinoma with ERBB2 exon 20 insertions: Comutations and immunogenomic features related to chemoimmunotherapy. Lung Cancer (2021) 160:50–8. doi: 10.1016/j.lungcan.2021.07.014 [DOI] [PubMed] [Google Scholar]
- 45. Azuma K, Ota K, Kawahara A, Hattori S, Iwama E, Harada T, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected non-small-cell lung cancer. Ann Oncol (2014) 25:1935–40. doi: 10.1093/annonc/mdu242 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this publish article/ Supplementary Material . Further inquiries can be directed to the corresponding author.