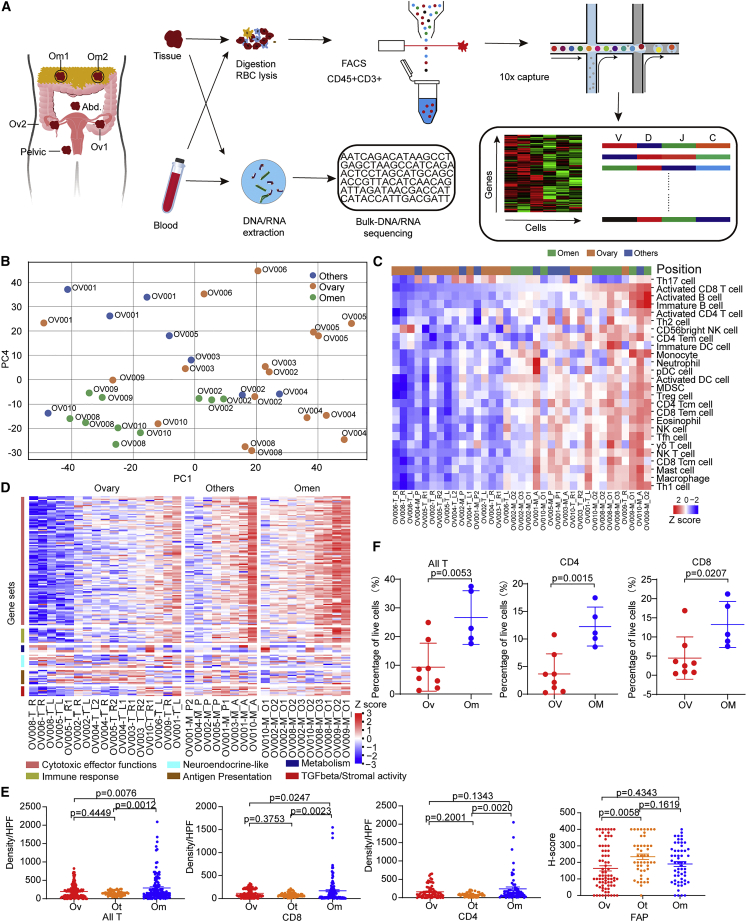
Figure 1.
Differential transcriptomic profiles across multiple sites in HGSOC
(A) Overview of the study design.
(B) The PCA plot of mRNA expression.
(C) The abundance of 28 immune cell types (identified by ssGSEA) is shown according to distinct locations.
(D) The gene expression of six immune-related pathways in tumors of different locations.
(E) Quantification of densities of CD4+ and CD8+ cells, and FAP H scores across three sites. Data represent mean ± SEM. p values were determined by Tukey’s multiple comparisons test.
(F) Quantification of all T (CD45+CD3+), CD4+ T (CD45+CD3+CD4+), and CD8+ (CD45+CD3+CD8+) T proportions in tumors from each sample, respectively. Ovarian samples = 8, omental samples = 5. Data represent mean ± SEM. p values were determined by Student’s t test.