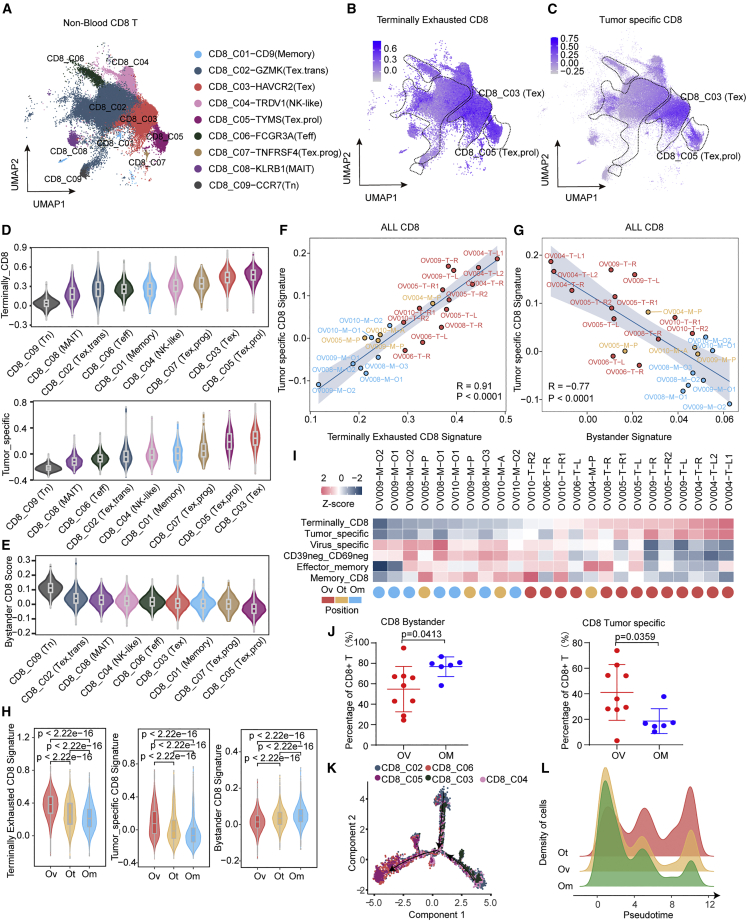
Figure 3.
Characterization of CD8+ tumor-infiltrating T cells in HGSOC
(A) UMAP of 94,424 single CD8+ tumor-infiltrating cells, showing the formation of 9 main clusters in tumor tissues.
(B and C) UMAP of CD8+ tumor-infiltrating cells colored according to gene signature scores, (B) terminally exhausted CD8+ signature, and (C) tumor-specific CD8+ signature.
(D) Violin plots showing the sorted gene signatures scores (up, terminally CD8+ signature score; down, tumor-specific signature score) across 9 CD8+ tumor-infiltrating cell clusters.
(E) Violin plots showing the sorted bystander CD8+ signature score across 9 CD8+ tumor-infiltrating cell clusters.
(F and G) Correlations between different gene signatures in all CD8+ tumor-infiltrating cells at the sample level, (F) terminally exhausted CD8+ signature score and tumor-specific CD8+ signature score, and (G) bystander signature score and tumor-specific CD8+ signature score, each color represents a different tumor site.
(H) Violin plots showing the gene signatures scores (left, terminally exhausted CD8+ signature score, middle, tumor specific CD8+ signature score, right, bystander CD8+ signature score) in CD8+ tumor-infiltrating cells from different positions, including Ov, Om, and Ot. wilcox.test.
(I) Heatmap showing multi gene signatures and sample positions information at the sample level, arranged from low to high by the terminally CD8+ signature score.
(J) Quantification of CD8+ bystander T (CD45+CD3+CD8+CD39−) and CD8+ tumor-specific T (CD45+CD3+CD8+CD39+) cells proportions in tumors from each sample, respectively. Ovarian samples = 8, omental samples = 5. Data represent mean ± SEM. p values were determined by Student’s t test.
(K) Potential developmental trajectory of CD8+ tumor-infiltrating cells inferred by Monocle2 based on gene expressions, each color represents a different cluster.
(L) Density plot showing the density patterns of cells from different tumor positions, including Ov, Om, and Ot along the pseudotime, each color represents a tumor site.