Figure 7.
Inherent TME characteristics contribute to spatial differences of TIL status
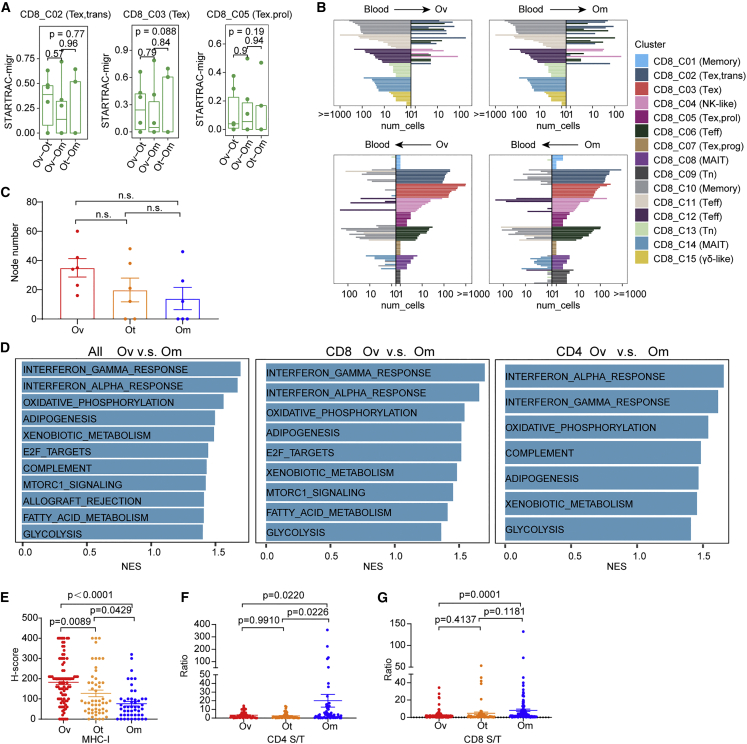
(A) Developmental migration of CD8+ tumor-infiltrating cells between every two of the three tumor sites quantified by pSTARTRAC-migr indices for each patient (n = 6), Kruskal-Wallis test.
(B) Top 10 shared clones of blood and tumor (bottom) being shared with tumor and blood, respectively, for each CD8 cluster. This analysis was performed in ovarian (Ov, left), and omental (Om, right) sites, respectively.
(C) The number of nodes of the network diagrams were counted and compared among tumor sites. Tukey’s multiple comparisons test.
(D) Enrichment plots from gene set enrichment analysis (GSEA) showing significantly differentially regulated pathways between ovarian and omental sites at the single-cell level in all CD3+ tumor-infiltrating T cell (left), CD8+ tumor-infiltrating cells (middle), or CD4+ tumor-infiltrating cells (right). NES, normalized enrichment score.
(E) Quantification of MHC class I H scores across three sites. Data represent mean ± SEM. p values were determined by ANOVA.
(F and G) Quantification of the ratio of densities of CD4+ (F) and CD8+ (G) cells in tumor and stromal area among tumor sites. Data represent mean ± SEM. p values were determined by Tukey’s multiple comparisons test.