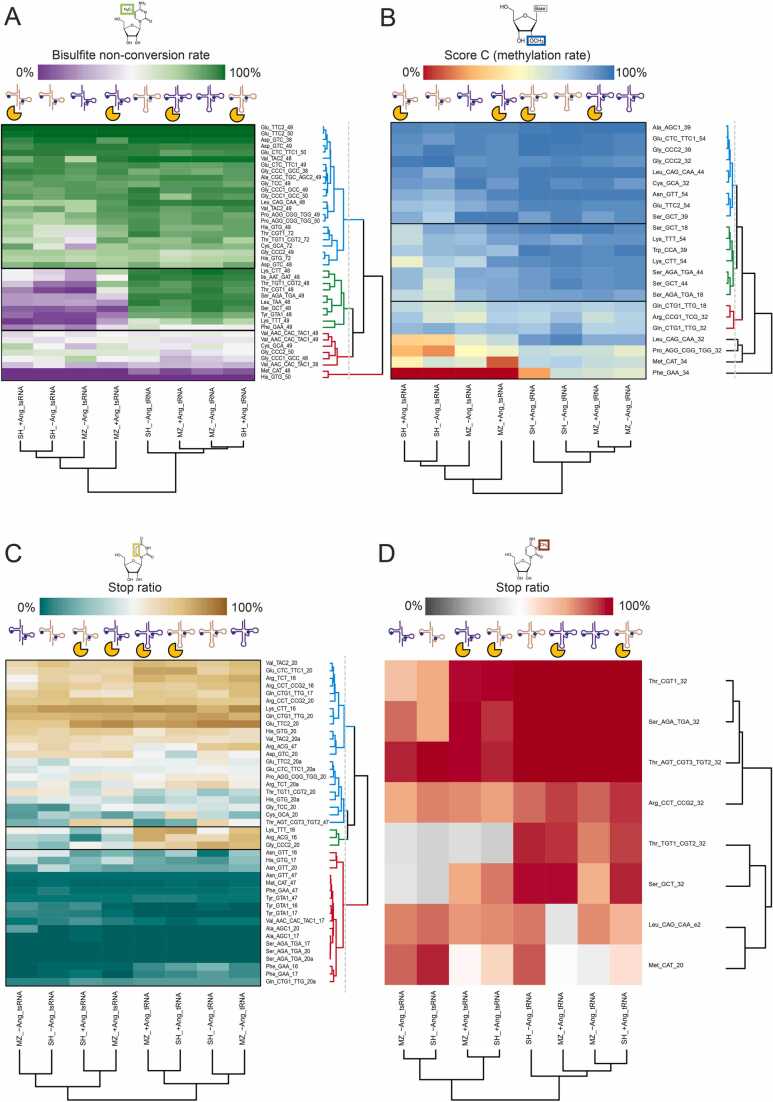
Fig. 7.
Heatmaps for the assessment of modification changes in individual modified sites in tRNAs/tsRNAs. Each heatmap displays one specific modification’s stoichiometry across the different samples (in X-axis) and the different sites retained for analysis (in Y-axis). Panel A displays m5C stoichiometry through bisulfite non-conversion rate (NonConv) from BisulfiteSeq, Panel B is for 2’-O-methylation through MethScore (also known as Score C) from RiboMethSeq and Panels C, D, and E are respectively for m7G, m3C and D though stop ratio from AlkAnilineSeq (see Material and Methods for further details). This stoichiometry is color-coded depending on the detection method used. Each sample and each site are hierarchically clustered based on the Euclidean distances between each subset, represented by dendrograms at the bottom for samples and on the right for sites. Dendrograms have been colored based on 3 main cluster properties: blue for sites with high stoichiometry and low variation, red for sites with low stoichiometry and low variation and green for sites with high stoichiometry variability. 4 sites remained unclassified for RiboMethSeq due to their unique behavior.