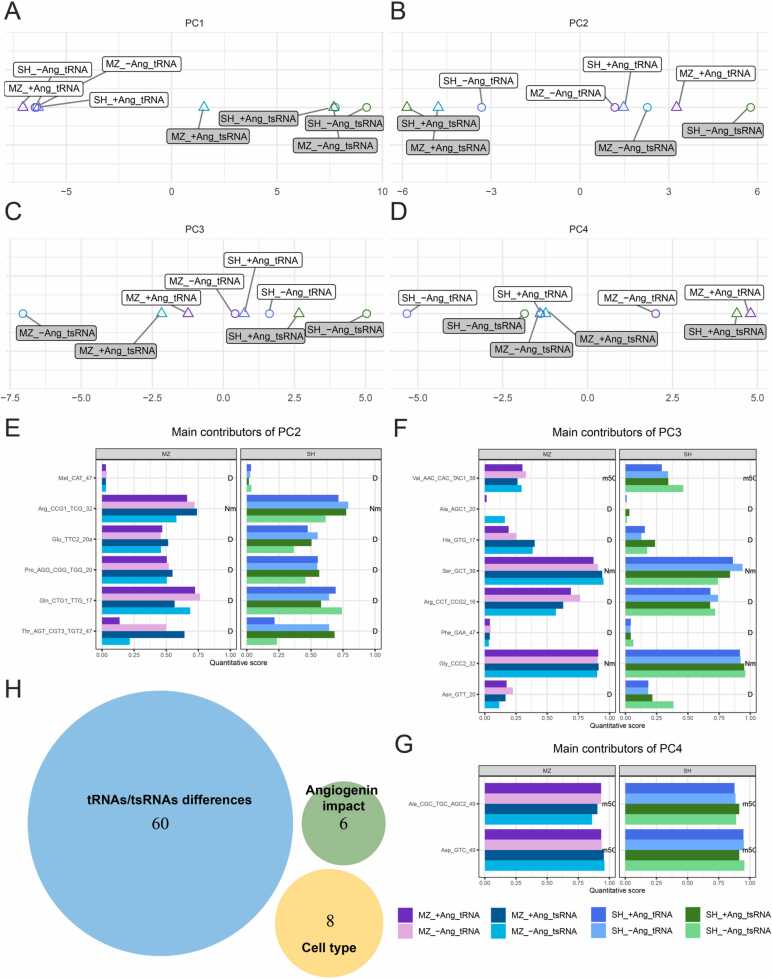
Fig. 9.
(A-D) Positions of datasets in 4 orthogonal vectors resulting from a PCA of all data points. Each principal component consists of linear combinations of modification data. The numbering PC1 through PC4 is according to the share of data that accounts for data variability: PC1 48.5%, PC2 15.4%, PC3 11.7% PC4 10.7%. (E-G) Barplots of the main contributors (eigenvectors) strongly correlating (absolute correlation coefficient > 0.70) with principal components 2, 3 and 4, respectively. (H) Venn diagram of the different contributors of the principal components and their interpretation. All PC1 main contributors have been labeled as “tRNAs/tsRNAs differences”, PC2 as “Angiogenin impact” and PC3 as “Cell type”. Main contributors to PC4 show negligible data variability and their information content is therefore negligible for the case at hand.