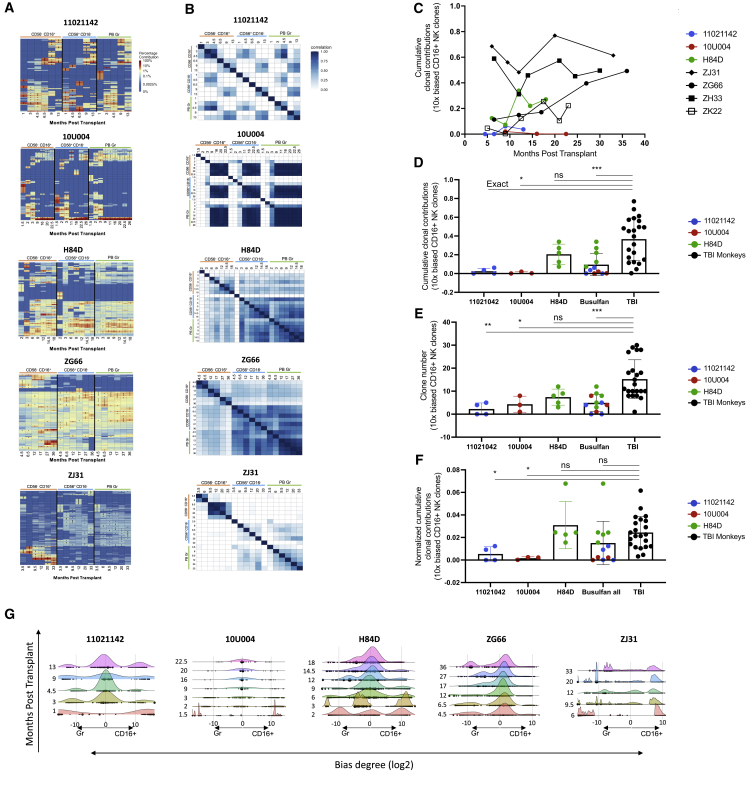
Figure 4.
Clonal patterns in NK cells
(A) Heatmaps depicting the contributions from the top 10 clones in CD16+ NK, CD56+ NK, and PB Gr samples over time post-transplantation for the three busulfan-conditioned animals and two TBI-conditioned animals (ZG66 and ZJ31). The heatmaps were generated as detailed as in Figure 2A. (B) Pairwise Pearson correlation coefficients between CD16+ NK, CD56+ NK, and PB Gr over time for each animal. The color scale for r values is shown on the right. (C) Total clonal contributions of the ≥10X biased and expanded CD16+ NK clones over time. A 10X biased clone was defined a top 30 contributing clone with a ≥10-fold expansion in fractional contribution to the CD16+ NK lineage compared with all other PB lineages analyzed (T, B, Mo, Gr, and CD56+) at the same time point. The three busulfan-conditioned animals and four TBI-conditioned animals with available samples (ZJ31, ZG66, ZH33, and ZK22) are shown. (D) Total clonal contributions of the 10X biased CD16+ NK clones at each time point post-engraftment comparing busulfan- and TBI-conditioned animals; 10X biased CD16+ NK clones defined as in (C) above. Unpaired t test applied with p < 0.05 considered as significant. (E) Number of 10X biased CD16+ NK clones contributing at each time point post-engraftment comparing busulfan- and TBI-conditioned animals; 10X biased CD16+ NK clones are defined as in (C) above. Unpaired t test applied with p < 0.05 considered as significant. (F) Normalized clonal contribution of each 10X biased CD16+ NK clone from busulfan and TBI-conditioned animals. Samples from different time points of individual busulfan-conditioned animals shown by animal or pooled together for comparison with the samples from four TBI-conditioned animals (ZJ31, ZG66, ZH33, and ZK22); 10X biased CD16+ NK clones are defined as in (C) above. Normalization: total clonal contributions of the 10X biased CD16+ NK clones from a sample divided by the clone numbers of the 10X biased CD16+ NK clones from the same sample. Unpaired t test applied with p < 0.05 considered as significant. (G) Ridge plot showing clonal bias between CD16+ NK and Gr lineages over time for busulfan- and TBI-conditioned animals. Ridges indicate the abundance-weighted density at the value of log-bias on the x axes, and dots indicate individual clones, sized by their overall abundance. Ridge plots stacked along the y axes correspond to the time point of each sample in months post-transplantation for each animal.