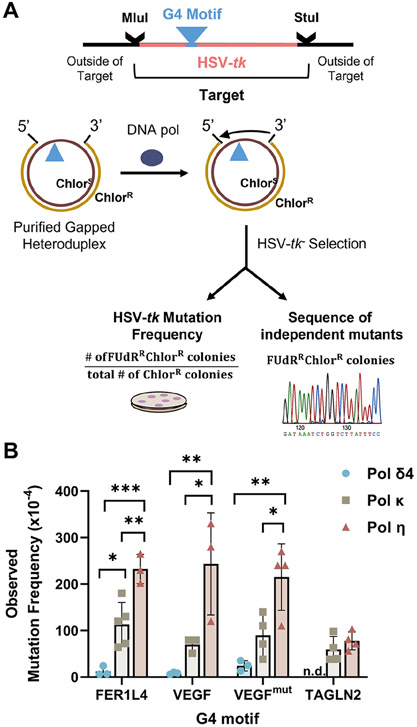
Figure 2. Human DNA Polymerase Error Rates Within the HSV-tk Assay.
(A) Schematic of assay. G4 motifs (▲) were inserted in-frame into the HSV-tk gene coding region between annotated restriction enzyme sites. The Mlu I to Stu I mutational target region contains the G4 motif and 97 bases of surrounding TK sequence. Gel-purified gapped heteroduplex DNA molecules were used as substrates by human polymerases η, κ, and δ4 (4-subunit). Reaction products that resulted in complete gap-filling were introduced into E.coli for HSV-tk − selection based on 5-fluoro-2’-deoxyuridine (FUdR) resistance. (B) Observed HSV-tk mutation frequency for each motif (Mean ± SD) and 3-4 independent reactions. Statistical analyses: One-Way ANOVA with Tukey test for multiple comparisons (pols δ4, κ, and η) for FER1L4, VEGF, VEGFmut motifs. Student’s t-test (two-tailed, unpaired) for TAGLN2. , *p<0.05, **p<0.01, ***p<0.001. n.d., not determined.