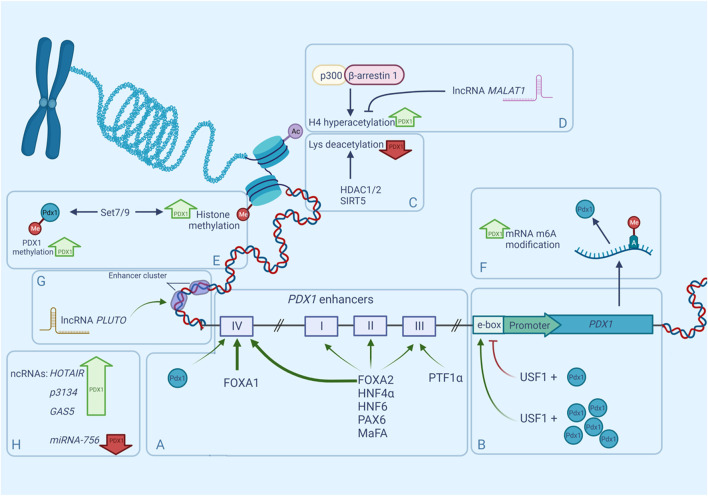
FIGURE 1.
Regulation of mammalian PDX1 gene by multiple factors. Expression of PDX1 is controlled by numerous epigenetic factors such as nucleosome positioning, histone methylation, histone acetylation, accessibility of enhancers and non-coding RNAs. PDX1 expression can be upregulated (bold green upward arrows) or downregulated (bold red downward arrows) by any of these factors. (A). Distant enhancers known as areas I-IV, participate in PDX1 transcription regulation due to their ability to bind transcription factors and boost gene expression. Same TFs can bind to multiple areas with different affinities, for example FOXA1/2 occupy area IV more efficiently (bold green arrows) than other areas (narrow green arrows). (B). regulation of PDX1 through upstream regulatory factors (USF) and E-box. (C). interaction of PDX1 and histone deacetylases. (D). interaction between β-arrestin-1, P300 and their role in PDX1 regulation. (E). PDX1 recruitment to Lys methyltransferase Set7/9. (F). RNA modification in PDX1 regulation. (G). (H). role of ncRNAs in PDX1 regulation.