Extended Data Fig. 10 |. piRNAi-induced transgenerational silencing.
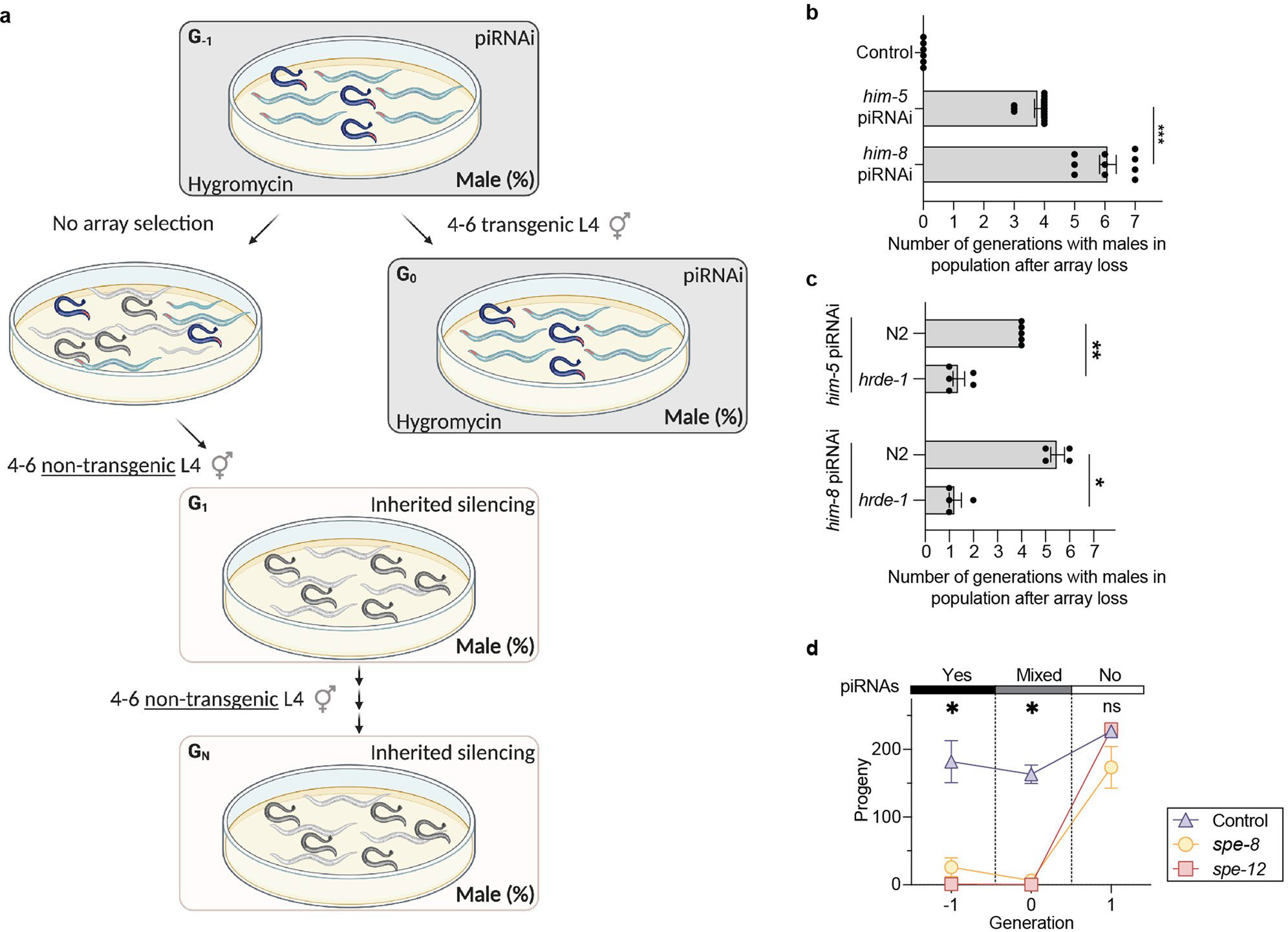
a. Schematic showing assay to determine him-5 and him-8 inherited silencing. To quantify the proportion of males generated from self-fertilization (as opposed to male cross progeny generated by mating), we picked four to six unmated L4 animals in every generation to new plates and scored their progeny for males (out of 100 animals). The assay has a ‘transition generation’ (G0) where a subset of animals are allowed to lose the extra-chromosomal piRNAi-array. Due to the shared germline cytoplasm, non-transgenic animals in the following G1 generation were exposed to sg-piRNAs in early embryonic development. The strains were not exposed to de novo sg-piRNAs generated during the L4 molt when sperm are made and female oocyte production is initiated. b. Duration of him-5 and him-8 transgenerational inheritance. We quantified transgenerational inheritance by counting the number of generations after losing the piRNA array before the fraction of males in the population was <= 1%. Data aggregated from panels Fig. 4bc and Supplementary Fig. 13. Kruskal-Wallis ANOVA, P=<0.0001, Dunn’s post hoc test *** P = 0.0007. c. Duration of transgenerational silencing in hrde-1 mutants. Two-tailed Mann-Whitney test, * P = 0.0286, ** P = 0.0079. d. Inherited silencing of piRNAi targeting spe-8 and spe-12 in a him-5(e1490) mutant background. Unmated L4 hermaphrodites were scored for viable progeny in the presence (solid black bar) or absence (solid white bar) of sg-piRNAs targeting spe-8 and spe-12, respectively. Control = him-5(e1490) animals with a randomized piRNAi cluster. Hygromycin was used to select transgenic animals, including controls, causing the increase in brood size of controls after the animals were grown on non-selective plates. Statistics: One-way ANOVA at each generation. * P = 0.0057 (generation −1), * P = 0.0127 (generation 0), ns P = 0.8312 (generation +1). Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Sample sizes (b) n = 6 (control), n = 14 (him-5), n = 8 (him-8), (c) him-5: n = 5 (N2), n = 5 (hrde-1), him-8: n = 4 (N2), n = 4 (hrde-1), (d) n = 2 (control), n = 2 (spe-8), n = 2 (spe-12) biologically independent transgenic strains.
