Figure 3 – piRNAi is specific and can be multiplexed.
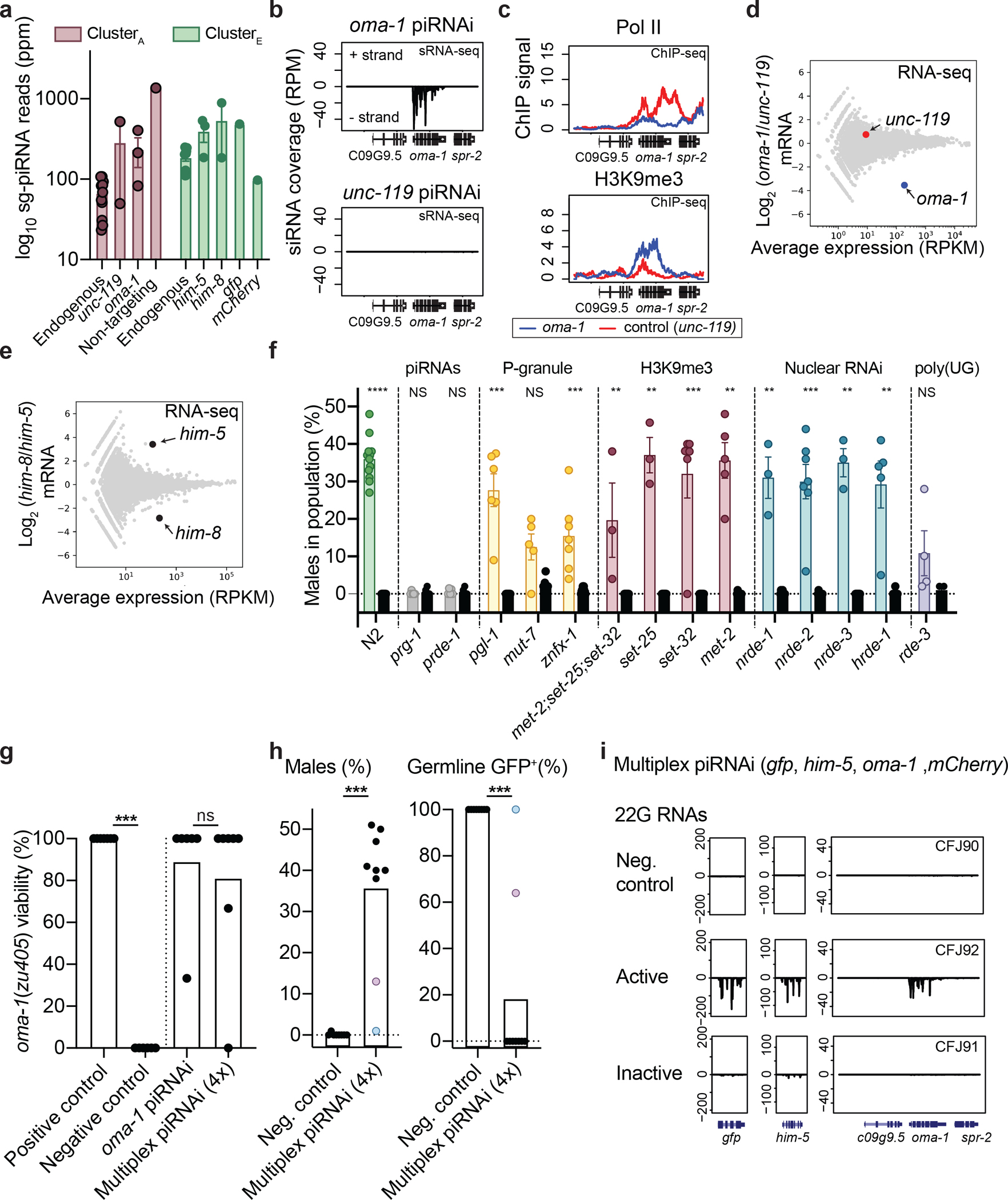
a. Total number of piRNA reads mapping to endogenous or sg-piRNAs from clusterA (centered on 21ur-199, purple) and the standard piRNA cluster (clusterE, centered on 21ur-1224, green). See also Supplementary Figures 4–5.
b. RNA-sequencing of secondary small RNAs (sRNA-seq) at the oma-1 locus in transgenic animals with piRNAi arrays targeting oma-1 or a control (unc-119).
c. Pol II (top) and H3K9me3 ChIP-seq (bottom) in strains with sg-piRNAs targeting oma-1 (blue) or unc-119 (red).
d. RNA sequencing (RNA-seq) of total RNA from young adult animals with piRNAi arrays targeting oma-1 (blue circle) or unc-119 (control, red circle).
e. Comparison of mRNA expression of a mixed population of young adult animals (hermaphrodites and males) with piRNAi against him-5 and him-8 (black circles).
f. Genetic requirements for silencing him-5 by piRNAi in N2 wildtype animals (green dots) or in mutant animals with primary effects on piRNAs (gray), P-granules (yellow), H3K9 methylation (red), nuclear RNAi (blue), and poly-UG tails (purple). Controls (black circles) are un-injected non-transgenic mutant strains. Mann-Whitney pairwise tests (injected vs un-injected). *** P < 0.0001, *** P < 0.001, ** P < 0.01, NS P > 0.05.
g. Multiplexed piRNAi-mediated silencing of a temperature-sensitive oma-1 allele using arrays with one (oma-1) or four (him-5, gfp, oma-1, and mCherry) piRNAi transgenes. Positive control = N2 injected with oma-1 piRNAi, negative control = oma-1(zu405ts). Mann-Whitney tests, P *** = 0.0006, ns = not significant (P = 0.853).
h. Multiplexed piRNAi against gfp and him-5 (in addition to oma-1 and mCherry) in transgenic Pmex-5::gfp animals. Neg. control = randomized sg-piRNAs. Purple and blue circles indicate matched data scored for males and GFP silencing in the same transgenic strain. Mann-Whitney test, P *** = 0.0001 (males) and *** = 0.0004 (GFP).
i. sRNA-seq from active (GFP negative and high male frequency) and inactive (GFP expression and no males) piRNAi arrays in Pmex-5::gfp transgenic animals (no mCherry transgene present in strain).
Data are presented as mean values +/− SEM with each data point corresponding to an independently derived transgenic strain. Negative controls = transgenic animals with randomized sg-piRNAs. Sense reads positive, antisense reads negative. Sample sizes (a) ClusterA: n = 11 (endogenous), n = 2 (unc-119), n = 3 (oma-1), n = 1 (random), ClusterA: n = 11 (endogenous), n = 3 (him-5), n = 2 (him-8), n = 1 (gfp), n = 1 (mCherry) small RNA libraries from biologically independent transgenic strains, (f) N2: n = 12 and n = 7, prg-1: n = 7 and n = 5, prde-1: n = 7 and n = 5, pgl-1: n = 6 and n = 7, mut-7: n = 5 and n = 7, znfx-1: n = 7 and n = 7, “met-2; set-25; set-32”: n = 5 and n = 7, set-25: n = 3 and n = 7, set-32: n = 3 and n = 7, met-2: n = 6 and n = 7, nrde-1: n = 3 and n = 7, nrde-2: n = 7 and n = 7, nrde-3: n = 3 and n = 7, hrde-1: n = 5 and n = 7, rde-3: n = 4 and n = 4 biologically independent transgenic strains (piRNAi and controls, respectively), (g) n = 7 (positive control), n = 6 (negative control), n = 6 (oma-1), n = 7 (multiplex) biologically independent transgenic strains, (h) n = 8 (negative control), n = 9 (multiplex) biologically independent transgenic strains.
