Abstract
Microfluidic tissue barrier models have emerged to address the lack of physiological fluid flow in conventional “open-well” Transwell™-like devices. However, microfluidic techniques have not achieved widespread usage in bioscience laboratories because they are not fully compatible with traditional experimental protocols. To advance barrier tissue research, there is a need for a platform that combines the key advantages of both conventional open-well and microfluidic systems. Here, we develop a plug-and-play flow module to introduce on-demand microfluidic flow capabilities to an open-well device that features a nanoporous membrane and live-cell imaging capabilities (“m-µSiM”). The magnetic latching assembly of our design enables bi-directional reconfiguration and allows users to conduct an experiment in an open-well format with established protocols and then add or remove microfluidic capabilities as desired. Our work also provides an experimentally-validated flow model to select flow conditions based on the experimental needs. As a proof-of-concept, we demonstrate flow-induced alignment of endothelial cells, the expression of shear-sensitive gene targets, and visualize the different phases of neutrophil transmigration across a chemically stimulated endothelial monolayer under flow conditions. With these experimental capabilities, we anticipate that both engineering and bioscience laboratories will adopt our reconfigurable design due to the compatibility with standard open-well protocols.
Keywords: tissue barrier, vascular barrier, modular, microfluidic, microphysiological system, organ on chip, tissue chip

Although Transwell-like platforms are the gold standard for creating tissue-barrier models in bioscience laboratories, they lack physiological fluid flows. In this work, a static open-well culture platform-featuring a 100 nm thick nanoporous membrane-is reconfigured between static and flow-enhanced culture modes to increase experimental flexibility. Using this platform, upregulation of shear-responsive genes and dynamics of neutrophil transmigration under flow are demonstrated.
1. Introduction
Endothelial-lined vascular barriers separate blood from the surrounding tissue and play a crucial role in maintaining homeostasis by regulating molecular permeability, recruiting immune cells, and preventing pathogen entry into the tissue compartment (Figure 1A).[1–3] Impaired barrier function or breakdown can have significant implications on human health, including stroke,[4] cancer,[5] cardiovascular disease,[4,6] and neurodegenerative disorders.[7,8] As a scalable companion to animal studies, in vitro culture models aim to replicate key aspects of the in vivo environment and are a popular method to study factors that affect barrier structure and function in a controlled manner.[2,8–11]
Figure 1.
(A) Schematic illustration of an in vivo vascular barrier. The pink domain represents fluid in the luminal side, and the green domain represents the abluminal side of the barrier. Fluid flow applies shear stress to endothelial cells and carries neutrophils to the barrier. During injury, neutrophils are recruited from the bloodstream in a coordinated process of rolling, arrest, crawling, and transmigration across the cell monolayer under continuous flow conditions in response to inflammatory factors. To mimic the vascular environment, in vitro models contain a membrane as a physical support for endothelial cells. (B) Comparison of the membrane characteristics and imaging quality between (i) conventional Transwell-like inserts and (ii) the open-well m-μSiM. Scale bars = 100 µm.
Barrier models, including the popular “open-well” Transwell™ - like format, aim to replicate the compartmentalization found in vivo by introducing a porous culture membrane separating the top and bottom media-filled compartments.[8,11] Cells are seeded directly onto the membrane that physically separates the compartments while permitting soluble factor communication between the cell populations.[8,11] This configuration is relevant to several barrier tissues, including the blood-brain barrier,[12,13] alveolar-capillary interface,[14,15] and glomerular barrier.[16] Studies to assess barrier structure and function are well-established and include fluorescence imaging of junctional proteins, molecular permeability measurements, and endpoint cell transmigration studies.[17–21] Although open-well devices are convenient and feature tried-and-true experimental protocols, they cannot introduce fluid flows to mimic the perfused in vivo environment that circulates immune cells and contributes to barrier maturation.[8,11,22] To address the lack of flow capabilities in conventional open-well assays, microfluidic platforms (e.g., microphysiological systems) that incorporate a porous culture membrane between the individually addressable top (luminal) and bottom (abluminal) fluidic channels have emerged.[23–40]
While microfluidic platforms offer distinct fluidic routing capabilities and feature dynamic control over biophysical and biochemical stimuli, they have not been widely adopted by bioscience laboratories where static, open-well assays remain the gold standard.[10,11,41] This issue can partly be attributed to incompatibilities between microfluidic and conventional experimental workflows. For example, initial cell seeding in microfluidic channels is carried out under flow conditions; cells travel along the fluidic path (e.g., the tubing and channel) and finally settle onto the culture surface.[11] Given the importance of the initial seeding density on barrier formation,[42] this stochastic seeding approach poses a challenge because precisely controlling the uniformity of cells on the membrane under flow can be difficult to replicate between experiments. Conversely, open-well seeding simply adds a known cell number directly to the culture membrane to achieve the desired density (i.e., cells.cm−2). Although many standard assays can now be performed in a microfluidic channel under controlled flow conditions (e.g., immunostaining, protein isolation, molecular permeability, and RNA extraction), the sample handling and experimental protocols are naturally more complex and less familiar than those used for static, open-well formats, and require significant optimization efforts.[2,11] An ideal system would unite the distinct advantages of open-well and microfluidic techniques and provide an opportunity to switch the configuration during an experiment based on the experimental requirements and user preference.
Here, we introduce a reconfigurable platform that combines the ease-of-use and standard protocols found in open-well formats with the controlled dynamic flow capabilities provided by microfluidics. Building from our open-well m-µSiM (modular microphysiological system enabled by a silicon membrane) platform described in a companion paper,[43] we use a tool-free magnetic latching approach to insert a microfluidic flow module onto an established endothelial barrier in the m-µSiM and introduce luminal flow for shear conditioning and leukocyte introduction. The m-µSiM features an ultrathin culture membrane with molecular level thinness (100 nm), high porosity (15%), and excellent live imaging capability that addresses problems with track-etched membranes materials used in commercial membrane-based culture systems (Figure 1B).[26,29–31] As shown in our companion paper, barriers can be established in the m-µSiM using familiar open-well protocols with excellent reproducibility across engineering and bioscience-focused laboratories.[43]
To highlight the flexible nature of our approach, we seed and establish an endothelial monolayer in the open-well format and reconfigure the platform into a fluidic system. Using particle image velocimetry (PIV), we experimentally validate the computational model of the fluid flow path and demonstrate the shear-induced alignment of endothelial cells. We then show compatibility with standard open-well RNA extraction techniques and confirm the upregulation of shear-sensitive gene targets. As an application, we introduce leukocytes to the endothelial barrier under flow conditions and visualize the distinct phases of cell recruitment, including rolling, arrest, crawling, and transmigration across the endothelial monolayer in response to abluminal N-Formylmethionine-leucyl-phenylalanine (fMLP) stimulation. By enabling the facile reconfiguration of the open-well m-µSiM into the fluidic mode, we aim to provide bioscience and engineering-focused researchers with a simple, modular workflow that provides on-demand flow capabilities.
2. Experimental Section
Device Fabrication
Open-well m-µSiM device:
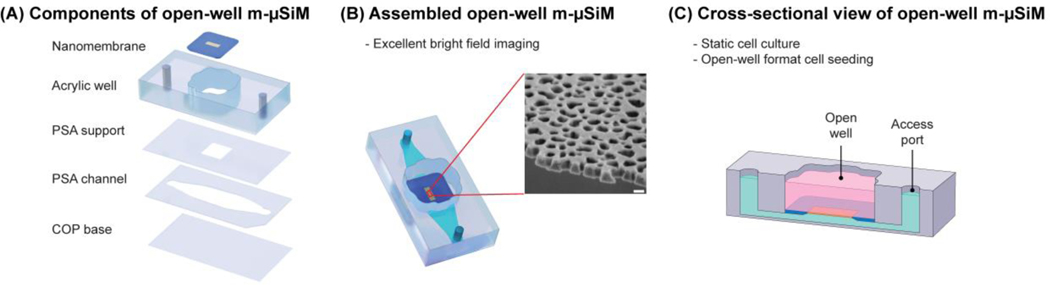
100 nm ultrathin nanoporous silicon nitride membranes with 15% porosity and 60 nm pore sizes were provided by SiMPore Inc. (Rochester, USA). The overall dimensions of the membrane chip, including the silicon support, were 5.4 5.4 0.3 mm with a central permeable window of 2 mm 0.7 mm 0.1 µm. m-µSiM components, including the transparent COP (cyclic olefin polymer) base, PSA (pressure-sensitive adhesive) channel and support layers, and acrylic layers, were purchased from Aline Inc. (Signal Hill, USA). The membrane and m-µSiM components were assembled in a layer-by-layer manner, as described in a companion paper (Figure 2A).[43] The porous region of the membrane (yellow) is surrounded by a silicon support (blue) (Figure 2B). As shown in Figure 2C, the membrane separates the two compartments of the m-µSiM into luminal and abluminal sides while allowing soluble factor communication between compartments to occur over length scales consistent with the ∼100 nm thick basement membrane found in vivo.[29–31]
Figure 2.
Schematic illustration of open-well m-μSiM. (A) Open-well m-μSiM consists of an acrylic top well and PSA microchannel separated by an ultrathin membrane. (B) Assembled view of the open-well m-µSiM showing the porous membrane (yellow) and silicon support (blue). The inset shows an SEM image of the 100 nm thick membrane. Scale bar = 100 nm. (C) A cross-sectional view of the open-well m-µSiM showing luminal (pink) and abluminal (green) compartments separated by the porous membrane.
Flow module:
A polydimethylsiloxane (PDMS) flow module (molded microchannel dimensions, w = 1.5 mm, h = 0.2 mm, l = 5 mm) was fabricated using standard soft lithography techniques.[44–46] Briefly, SU-8 3050 photoresist (Kayaku Advanced Materials, USA) was spin-coated onto a 100 mm diameter silicon wafer (University Wafers, USA), followed by a soft bake at 95 °C for 45 min. The photoresist was then exposed to UV light (250 mJ.cm−2) through a high-resolution printed photomask, baked for 5 minutes at 95 °C, and then developed in SU-8 developer until the features were resolved. The wafer was rinsed with IPA and dried using a stream of air. To fabricate flow modules with a clover shape and desired overall thickness (3.3 mm), an acrylic divider with a thickness of 3.3 mm was laser cut and attached to the wafer with PSA (MP467, 3M, USA) (Figure S1A). Degassed PDMS prepolymer (10:1 base to catalyst ratio) (Sylgard 184, USA) was poured over the mold and a ruler was used to level the upper surface before curing for 1 h at 70 °C on a hotplate. The flow modules were extracted from the mold and access ports were cored using a 1 mm biopsy punch (World Precision Instruments, USA).
Housings:
Upper and lower housings were fabricated by laser-cutting acrylic sheets with a thickness of 2 and 3.5 mm, respectively. The lower housing was bonded to a coverslip using PSA. Nickel-plated magnets with a diameter of 4.75 mm and pull force of 0.34 Kg (K&J Magnetics, USA) were press-fit into the housings.[47,48]
Patterning stencil:
To create stencils with cone-shaped wells (h = 3.3 mm, taper angle = 57º), a custom aluminum mold was milled using a CNC machine and then polished. An acrylic divider with a thickness of 3.3 mm was laser cut and attached to the mold with PSA to obtain stencils with a clover-shaped footprint (Figure S1B). Degassed PDMS prepolymer (10:1 base to catalyst ratio) was poured over the mold and cured for 1 h at 70 °C on a hotplate.
Flow circuit:
A custom-made flow circuit (Figure S2) was designed and fabricated using sample collection vials, Gauge 21 NT dispensing tips (Jensen Global, USA), 0.22 µm PVDF syringe Filters (Perkin Elmer, USA), and 0.5×0.86 Micro Flow tubes (Langer Instruments, USA). A peristaltic pump (Langer Instruments, USA) was used for flow circulation.
Burst Pressure and Leakage Testing
In the burst pressure test, dyed water was introduced to the luminal side of the device using pressurized air. The device outlet was then closed using a 4-Way Stopcock (Qosina, USA). The supply pressure was increased gradually until membrane damage was observed. For the leakage test, dyed water was introduced into the device using a syringe pump and the flow rate was increased sequentially until leakage was detected.
Simulation
A steady-state 3D simulation of the fluid domain was performed using laminar flow physics in COMSOL Multiphysics®. Constant inlet velocity corresponding to a range of flow rates (Q=10, 100, 200, 500, and 1000 μl.min−1) was applied to the inlet, while the outlet boundary condition of pressure P=0 was applied to the outlet. Wall boundary conditions were applied to all other surfaces. The physics-controlled mesh was used and a mesh independence study was conducted. For the waveform analysis, a sinusoidal wave with 1Hz frequency was applied as the inlet boundary condition and a time-dependent study with t=0:50:500 ms was used. To improve the accuracy of the solutions, a second-order discretization was utilized.
Particle Image Velocimetry (PIV)
To validate the COMSOL model, experimental fluid flow analysis was carried out using particle image velocimetry on a Nikon Eclipse TE2000-S microscope equipped with a pulsed laser (TSI, MN, USA) (Figure S3). For this purpose, 5 μm fluorescent polystyrene latex particles (Magsphere, CA, USA) in deionized (DI) water were injected at defined flow rates (Q=10, 100, 200, 500, and 1000 µL.min−1) into the chip using a syringe pump. The PIV measurement was carried out on three independent devices and was repeated three times at each flow rate. All measurements were conducted in the middle of the channels (z=100 µm). For each measurement, 50 pairs of images were captured. The time interval between image pairs was varied from 100 to 1000 μs depending on the flow rate. The velocity was determined for each image pair using cross-correlation algorithms, and the results of the 50 instantaneous measurements were used to calculate the mean velocity for each replicate device. All data capture and analysis were performed using TSI Insight 4G™ software (TSI, MN, USA).
Cell Culture
Human Umbilical Vein Endothelial Cells (HUVEC) (Thermo Fisher Scientific, USA) were cultured in EBM™-2 Basal Medium (Lonza, USA) supplemented with EGM™-2 Endothelial Cell Growth Medium-2 BulletKit™ (Lonza, USA). HUVECs were used between passages 3–6. Cells were cultured in T25 flasks and were enzymatically dissociated using TrypLE (Thermo Fisher Scientific, USA) for 3 minutes and centrifuged at 150 G for 5 minutes, and re-suspended. To improve cell attachment, the m-µSiM membrane was coated with 5 µg.cm−2 Fibronectin (Corning, USA) for 1 hour at room temperature. A patterning stencil was inserted in the m-µSiM well and cell media was added to the well and bottom channel of the device. Next, cells were seeded at a density of 40,000 cells.cm−2 into the well and incubated for 24 hours at 37o C and 5% CO2. For the conventional open-well device, ThinCert™ transparent polyethylene terephthalate inserts (Greiner Bio-One, USA) were purchased, and a similar cell seeding procedure was carried out. To introduce shear flow to the cells, the m-µSiM was reconfigured into a fluidic system and cells were subjected to 10.7 dynes.cm−2 at a flow rate of 580 µL.min−1 for an additional 24 hours. Before use, the flow module, housings, and flow circuit were sterilized via exposure to UV light for 30 min. Next, PBS 1X (VWR, USA) was run through the flow circuit for 2 min. Then, reservoirs and tubes were filled with cell media, and the assembled device was connected to the flow circuit using Gauge 21 dispensing tips (Jensen Global, USA).
RNA Extraction and Gene Expression Analysis
After shear stimulation, the flow module was removed and the membranes were washed twice with PBS. To lyse the cells, 100 μl of TRI-Reagent® (ThermoFisher Scientific, USA) was added to each device well and incubated for 5 minutes at room temperature. The lysate was then transferred into 1.5 ml RNase-free tubes and 1 ml of chloroform was added to each tube. After 5 minutes of incubation at room temperature with periodic mixing/shaking, the tubes were centrifuged at 16000 g and 4 °C for 15 minutes. The upper aqueous phase was collected and transferred into a new tube, and an equal volume of ice-cold isopropanol was added. The solutions were precipitated at −20 °C for 30 minutes and were then centrifuged at 16000 g and 4 °C for 30 minutes. RNA pellets were collected and washed sequentially with 70% and 100% ethanol. The solutions were air-dried for 5 minutes and resuspended in 11 ul of RNase-free water. RNA yield was measured using NanoPhotometer® N60/N50 (Implen, Germany). High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, USA) was used to reverse transcribe 1 μg of RNA into Complementary DNA (cDNA) in a 20 μl volume reaction. Quantitative PCR with 10 ng cDNA per reaction was performed using TaqMan™ Fast Advanced Master Mix (Applied Biosystems, USA). The reaction was run in a QuantStudio™ 3 RT-qPCR system (ThermoFisher Scientific, USA) to evaluate changes in the expression of the following genes: eNOS, KLF2, and GAPDH as the housekeeping gene (Hs01574659_m1, Hs00360439_g1, Hs02786624_g1, ThermoFisher Scientific, USA). The following amplification conditions were used: 2 minutes at 50 °C, followed by 2 minutes at 95°C, then 40 cycles of 95 °C for 1 s and 60 °C for 20 s. Two technical replicates were used for each sample and the ΔΔCT method was used to calculate the relative expression level considering GAPDH as a reference gene.
Neutrophil Isolation and Transmigration
Polymorphonuclear neutrophils were isolated using a protocol described in Salminen et al., which was approved by the Research Subjects Review Board at the University of Rochester.[29,49] Briefly, venous whole blood from consenting healthy donors was collected into sodium heparin-coated vacuum tubes and cooled to room temperature. Blood samples were carefully layered on top of equal volumes of 1-Step Polymorph solution (Accurate Chemical & Scientific Co., Westbury, NY) and centrifuged at 500g for 30 minutes. Post centrifugation, all density separation components (except for neutrophil-rich layers) were discarded. Neutrophils were extracted into 15 ml conical tubes and washed twice in 10 mL of wash buffer (calcium and magnesium-free Hank’s balanced salt solution, 10 mM HEPES Sodium Salt, and 5 mg.mL−1 bovine serum albumin) via pelleting at 350g for 10 minutes, supernatant extraction, and subsequent resuspension. The washed neutrophil-rich solution was then depleted of red blood cells via RBC lysis (4.5 mL of 1/6th x PBS for one minute followed by 1.5 mL of 4x PBS for balancing) before one final wash with 10 mL of wash buffer. After the final wash, the neutrophil-rich pellet was resuspended in 1 mL of wash buffer, transferred to a 1.5 mL conical tube, and left in a rotating stand to prevent neutrophils from settling.
Neutrophils were introduced to the system for transmigration studies once a fully confluent monolayer of endothelial cells was formed. Isolated neutrophils from the resuspended stock were aliquoted into MCDB-131 complete medium (VEC Technologies, Rensselaer, NY) at 3 million cells.mL−1 and introduced into the top channel following a flow-stepping process of 10, 1, 0, 1, 10 µL.min−1 (30 seconds each until 10 µL.min−1 is reached again and maintained for 30 minutes) using a syringe pump. 10 nM fMLP (N-Formylmethionyl-leucyl-phenylalanine) (Sigma Aldrich, St. Louis, MO) was incorporated into MCDB-131 media and introduced to the bottom channel of the device to serve as a neutrophil chemoattractant. Phase-contrast microscopy (Nikon Eclipse Ti2, Tokyo, Japan) at 40x was utilized to record a video (Andor Zyla sCMOS, Belfast, United Kingdom) at 20 Hz of rolling, arrest, crawling, and transmigration events in the m-µSiM device. All neutrophil experiments were performed within 5 hours of blood draw.
Flow Cytometry
Neutrophils were aliquoted into negative control, positive control, and flow module groups before antibody staining and flow cytometry. All aliquots consisted of 1 mL of neutrophil-rich fluid and were deposited into 1.5 mL conical tubes. Negative control neutrophils were suspended in wash buffer at a concentration of 900k cells.mL-1. Positive control neutrophils were incorporated into MCDB-131 Complete Media at a concentration of 900k cells.mL−1 and supplemented with 10 ng.mL−1 IL-8 (rh CXCL8/IL-8, R&D Systems, Minneapolis, MN) for 15 minutes before rinsing and stained. Both control groups were left on a rotating stand to prevent neutrophils from settling. For the flow module group, neutrophil-rich fluid from a non-agitated reservoir containing 3 million cells.mL−1 (suspended in MCDB-131 Complete) was pulled through the m-µSiM device at a flow rate of 10 µL.min−1 using a syringe pump (Chemyx Fusion 200, Stafford, TX) over 40 minutes. To remove potential effects of syringe geometry and associated shear stresses on neutrophil activation, 152 mm of 0.8 mm inner diameter silicone tubing (Model 95802–01, Cole Parmer, Vernon Hills, IL), was placed between the syringe pump and flow module. This length held a volume of ∼300 µL neutrophil-rich media, which resulted in 900k cells after accounting for ‘loss’ from the volume of the fluidic circuit/device, and media from this section of tubing was exclusively used for analyzing flow module neutrophil activation. The samples were collected in separate 1.5 mL conical tubes, pelleted, and resuspended in 400 µL buffer (Flow Cytometry 1x Staining Buffer, R&D Systems, Minneapolis, MN). The 400 µL suspension for each experimental condition was then distributed into four 100 µL aliquots in 1.5 mL conical tubes representing the unstained control, isotype control, and L-selectin groups. Staining was performed according to manufacturer protocol using antibodies conjugated to Alexa Fluor 488 (R&D Systems, Minneapolis, MN). Flow cytometry was performed post live staining on a Guava EasyCyte benchtop machine. Sampling was set at 3000 gated events and data gathered was analyzed in FCS Express, GraphPad PRISM, and Wolfram Mathematica.
To assess neutrophil activation rates in the flow module, a histogram distance measurement (Jaccard Index) was calculated utilizing green fluorescence intensity data.[50] As L-selectin is cleared from an activated neutrophil membrane surface via ectodomain shedding, decreased expression of L-selectin in an activated neutrophil population will result in greater rates of non-specific binding for anti-L-selectin staining antibodies.[51–53] Activated neutrophil populations displayed higher degrees of overlap between respective L-selectin expression histograms and isotype control histograms. Higher degrees of neutrophil activation corresponded to higher Jaccard Index values, which were calculated for each experimental group.
Immunostaining and Imaging
For actin and nuclei staining, cells were fixed in 4% paraformaldehyde (Fisher Scientific, USA) for 15 min. Then, cells were permeabilized in Triton X-100 (0.1%) for 10 minutes and washed with PBS Tween-20 (PBST). Next, cells were blocked in 40 mg.mL−1 BSA (Alfa Aesar, USA) for 30 minutes at room temperature. Cells were labeled with Hoechst 33342 (300 nM) (Molecular Probes, USA) for 10 minutes and AlexaFluor 488 conjugated phalloidin (ThermoFisher Scientific, USA) (1:400) for 15 minutes to visualize nuclei and actin fibers, respectively. Finally, cells were washed with PBS Tween-20 and stored in PBS. For viability assays, the LIVE/DEAD™ Cell Imaging Kit (Thermo Fisher Scientific, USA) was used based on the vendor’s protocol. In the case of imaging cells on the silicon support of the membrane, a coverslip was placed on the m-µSiM and the device was flipped and imaged from top to bottom. Phase and fluorescence imaging of cell samples was performed on an Olympus IX-81 inverted microscope with CellSens software (Olympus, Japan). All image collection settings were consistent across experimental sets to allow comparison.
Customization of m-µSiM and introduction of aligned ECM fibers
A customized “segmented” channel geometry (wide segment: L = 5mm, W = 4mm, narrow segment: L = 5 mm, W = 1 mm) was fabricated by laser-cutting a 130 µm PSA sheet. During assembly of the m-µSiM, the standard bottom channel was replaced with the customized channel. Atelo-bovine collagen type-1 (Nutragen, Advanced BioMatrix, CA, USA) with a final concentration of 2.5 mg.mL−1 was prepared as described previously.[45,54] The diluted solution was injected into the custom-designed bottom channel of the m-µSiM through a Gauge 20 IT dispensing tip at a flow rate of 200 µl.min−1 using a syringe pump. The device was maintained at 37°C in an incubator for one hour to induce collagen polymerization. Collagen fibers were imaged using a Leica SP5 confocal microscope (Leica Microsystems, Germany) with a 40X water objective (HCX PL APO 40x/1.1 W) at 1.7x optical zoom. At each location, stacks with a thickness of 2 μm comprising 13 slices were captured.
Image analysis
For cell alignment quantification, the Analyze Particles module in FIJI (NIH, USA) was used, and then results were plotted as radar graphs using MATLAB CircHist plugin.[55] For collagen fiber quantification, the image stacks were projected onto a maximum projection plane in FIJI. LOCI CT-FIRE was then used to identify collagen fibers in the projected confocal reflectance microscopy (CRM) image and calculate the coefficient of alignment (CoA).[45,54,56]
Statistical Analysis
All experiments were independently performed in triplicate and the results were reported as mean ± SD unless otherwise stated. Experimental groups and controls were performed in parallel. For the gene expression analysis, in addition to three independent biological replicates, two technical replicates were used for each qPCR run. In the relative gene expression analysis, ΔΔCT values were transformed into log10 for the statistical analysis. For both gene expression analysis and the neutrophil activation assay, normal distributions were confirmed, and an unpaired parametric two-tailed t-test was conducted. For all statistical analyses, p<0.05 was considered to be significant. GraphPad Prism 9.3.1 was used for analysis and plotting.
3. Results
3.1. Reconfiguration of the static open-well m-µSiM into a flow-enhanced culture device
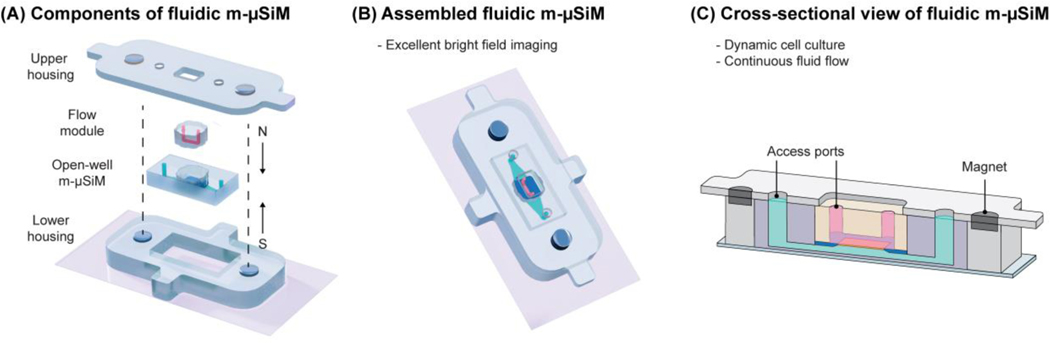
In our companion paper, we demonstrated that the open-well m-µSiM platform could support the formation and maturation of a blood-brain barrier mimetic using static cultures of iPSC-derived endothelial cells in different laboratory environments (engineering and bioscience laboratories) using mass-produced components and a common culture protocol.[43] To expand the experimental capabilities of the m-µSiM platform, we demonstrate that the open-well format can be reconfigured into a fluidic device using magnetic assembly.[47,48,54] As shown in Figure 3A, the core m-µSiM is first placed into a cavity defined by a lower housing and a coverslip. A molded “flow module” containing a lithographically defined channel and access ports (Figure S4) is added to the open-well reservoir. The flow module is sealed against the silicon support layer using magnetic “latching“ produced by the attraction force from magnets embedded in the top and bottom housing layers (Figures 3B, C, Video S1). The matching “clover” shaped geometry of the flow module and reservoir ensured that the microchannel features were positioned onto the silicon support without damaging the porous nanomembrane.
Figure 3.
Schematic illustration of modular fluidic m-μSiM. (A) The open-well m-μSiM is reconfigured into the fluidic m-μSiM using magnetic sealing of a flow module by upper and lower housings with embedded magnets. (B) Assembled view of the fluidic m-µSiM showing fluid in the flow module channel above (pink) and below (green) the membrane. (C) Cross-sectional view of the reconfigured fluidic m-µSiM with the flow path over the membrane identified.
To characterize the magnetic latching, we conducted a burst pressure test and showed that our system can tolerate dead-ended pressures up to 38.8 ± 2.4 KPa (Video S2), which is much higher than the operating pressure in many cell culture applications.[57] Furthermore, we showed that the system does not leak when using flow rates up to 4000 µl.min−1, which is equivalent to 74 dynes.cm−2 shear stress at the culture region (Video S3).
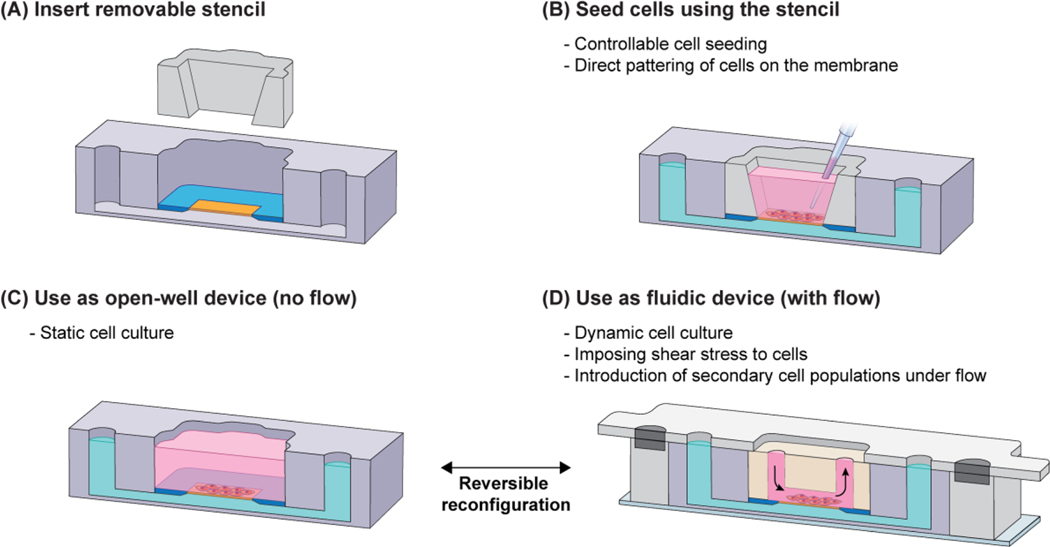
Although it is not a typical consideration for conventional static open-well platforms, the cell seeding approach must be carefully considered when developing a platform that can be reconfigured between open-well and fluidic modes. For example, as shown in Figure 4A, during cell seeding into the open-well m-µSiM, the HUVECs settled onto the membrane and surrounding silicon support and formed a continuous monolayer. When the flow module was magnetically sealed, cells outside of the microchannel boundary compressed and appeared non-viable (red), while cells inside the channel remained viable (green). Damage to the monolayer around the periphery of the channel may be problematic because the overall integrity of the barrier could be compromised and confound experimental findings. To alleviate such concerns, we designed a removable stencil that fits within the m-µSiM well and defines a patterning region where cells settle preferentially onto the membrane surface. Although there is an additional step in the workflow, cells are localized to a region within the microchannel boundary and cell damage does not occur after reconfiguration (Figure 4B).
Figure 4.
Comparison of open-well cell seeding vs stencil-based cell seeding. (A) Open-well format cell seeding results in cell settlement all over the well surface and consequent establishment of a monolayer on the membrane as well as its surrounding silicon chip. In this scenario, cells on the membrane’s surroundings are damaged upon inserting the flow module. (B) Whereas, in stencil-based seeding, cells are positioned selectively; hence, the monolayer only forms on the membrane surface and there are no cells in the surrounding of the membrane to be damaged upon inserting the flow module. Scale bars = 200 µm.
The placement of the stencil within the m-µSiM is compatible with conventional cell seeding protocols and is the first step in the experimental workflow (Figure 5A, B). After patterning and formation of a confluent cell monolayer, the user has two options: (i) for static cultures, the device can be used as-is in the open-well platform (Figure 5C), (ii) for flow experiments, the device can be reconfigured into the fluidic mode as shown in Figure 5D. The reversible magnetic latching mechanism supports on-demand reconfiguration between the open-well and fluidic formats (Video S1). For example, after flow stimulation, the device can be reconfigured to the open-well format and the user has the option to perform assays including immunostaining, RNA extraction, and barrier permeability measurements using standard experimental protocols as described here and in the companion paper.[43] The user also has the option to keep the channel in place and use microfluidic assay protocols if desired.[58,59]
Figure 5.
Experimental workflow for the fluidic m-µSiM. (A) Before cell seeding, a removable stencil is inserted into the m-µSiM well; then, (B) cells can be directly patterned on the membrane to prevent the distribution of cells on the membrane surrounding and consequent cell damage upon insertion of the flow module. (C) After the monolayer is established, the stencil can be kept or removed and the device can be used for static cell culture in the open-well format. (D) If desired, the user can reconfigure the device into a fluidic mode by inserting the flow module in the m-µSiM well and sealing it magnetically. After the flow experiment, the housings and flow module can be removed to reconfigure the system to the open-well format with direct access to the cells. The pink and green domains represent fluid in the vessel side and the tissue side of the barrier, respectively.
3.2. Computational modeling and experimental validation of flow characteristics in the fluidic m-µSiM
Flow-induced shear stress is a well-known biophysical cue that influences the structure and function of vascular barriers (e.g., gene expression, endothelial alignment, barrier integrity, and permeability).[8,60] To characterize the flow within the device, we developed a COMSOL model and experimentally validated the simulation using particle image velocimetry (PIV). Our results showed that the simulated and measured velocities matched within 10% (n=3), with excellent agreement over the tested range (linear regression, R2=0.99) (Figure 6A, B). Since there was a match between the experimental and simulated velocities, COMSOL results were used as a guide to calculate fluid-induced shear stresses at the cell monolayer and identify maximum shear stress values along the flow path (Table S1).
Figure 6.
(A) Schematic of the m-µSiM with attached flow module. The flow path is shown in pink. The velocities from the simulation and PIV were obtained from the mid-plane of the microchannel at z = 100 µm (dashed line) at Q = 10, 100, 200, 500, 1000 µL.min-1. (B) The comparison of velocities from the simulation and PIV measurements (R2 = 0.99, n = 3) shows excellent agreement. (C) A top-down view confirms that there is a uniform shear stress distribution over the culture region (denoted by the dashed rectangular lines).
Endothelial cells are exposed to fluid-induced shear stresses in vivo and their elongation along the flow direction results in enhanced barrier function.[8,30] With ∼10 dynes.cm−2 recognized as the typical threshold required for the alignment of endothelial cells,[30,61,62] we performed a parametric study using COMSOL to determine the flow rate required to produce a wall shear stress of 10 dynes.cm−2 in our flow module. Based on the simulation results, we selected Q = 580 µL.min−1 as the inlet flow rate, corresponding to 10.7 dynes.cm−2 shear stress at the membrane surface. The top-down shear stress distribution at the endothelial surface is shown in (Figure 6C). Although there are shear stress variations along the flow path resulting from the changes in geometry, the shear stress is uniform over the culture membrane.
This work is focused on providing steady laminar flow with minimal fluctuations, since that is a common experimental use case. For long-term studies using a peristaltic pump, we included flexible tubing and a damping reservoir to reduce unwanted pulsation (Figure S2). To further show the compatibility of our design with more complex waveforms, we used our experimentally validated COMSOL model to simulate a time-varying inlet flow (Q=Q0 sin (2πt) with Q0=580 µL.min−1 & t=0:50:500 ms). As shown in Figure S5A,B and Video S4, the time-varying input flow rate produced a uniform shear stress distribution over the culture region, with a magnitude varying sinusoidally from 0 – 10.7 dynes.cm-2. Since the Womersley number (W0) in the flow module is relatively small (W0 = 0.53 at 1Hz), the parabolic velocity profile has sufficient time to develop during each cycle.[63] Based on the channel geometry, our system can support waveforms up to 4 Hz without flow oscillation effects (i.e., W0 < = 1). Taken together, our results show that our flow module can support a wide range of input flow profiles.
To confirm that the predicted flow rate was sufficient to align endothelial cells, the open-well m-µSiM was reconfigured to the fluidic m-µSiM after initial monolayer formation, and cells were exposed to 10.7 dynes.cm−2 of shear stress (i.e., Q = 580 µL.min−1). Consistent with the literature,[30,61,62] shear-stimulation resulted in cell alignment (89±4.9 %) along the flow direction (Figure 7A), while cells in the static culture control (no flow) remained randomly orientated (Figure 7B). We have obtained similar shear-induced alignment of HUVECs using the reconfigurable approach between our engineering laboratories (Rochester Institute of Technology and the University of Rochester) with common components and protocols showing excellent inter-laboratory reproducibility using the flow module.
Figure 7.
Comparison of endothelial cells cultured in open-well and fluidic configurations of the m-μSiM. First, cells were seeded and maintained in open-well format for 24 hours to establish a confluent monolayer in m-μSiM devices. Then, one of the devices was reconfigured into the fluidic mode to induce shear alignment. (A) Cells cultured under dynamic conditions showed alignment along the flow direction due to continuous exposure to 10.7 dynes.cm−2 shear stress at a flow rate of 580 μL.min-1. The inset shows actin and nuclei of aligned cells in green and blue, respectively. Scale bar = 100 µm. (B) Cells cultured under static culture showed no alignment. Radar plots quantify cell alignment with respect to the flow direction (x-axis). The length of each bar represents the number of cells in the corresponding direction. Scale bar = 100 µm. (C) Comparison of the relative expression of KLF2 and eNOS between cells cultured under flow and cells cultured in open-well m-μSiM under static condition (**p < 0.01, n=3).
3.3. Compatibility of the platform with standard open-well protocols
Studies have shown that shear-stimulation of endothelial cells results in the upregulation of the shear-sensitive transcription factor Kruppel-like factor 2 (KLF2).[64,65] KLF2 plays a critical role in the regulation of endothelial activity such as the regulation of vascular tone, anti-inflammatory responses, and antithrombotic functions.[64] KLF2 is also responsible for the upregulation of endothelial nitric oxide synthase (eNOS) that promotes nitric oxide production and plays an atheroprotective function in healthy blood vessels.[65,66] To demonstrate that HUVECs exhibit expected behaviors in our system, we evaluated differences in KLF2 and eNOS gene expression in shear stimulated and static conditions.
We first established a barrier in open-well format, then reconfigured the device and stimulated cells with flow, and then reconfigured the device to open-well. RNA extraction was carried out using standard open-well protocols. Our results indicated that flow-stimulated cells (24 hours shear stress = 10.7 dyne cm−2) expressed 6.7x and 3.4x and higher levels of KLF2 and eNOS, respectively (Figure 7C). These readouts emphasize the importance of incorporating flow to mimic physiological conditions in barrier tissue models. This analysis also demonstrates the versatility of our platform for conducting downstream analysis using standard protocols designed for open-well devices. To further show the compatibility of our platform with standard protocols, we performed multi-step immunostaining on aligned cells. We removed the flow module after cell alignment and stained for actin and DAPI in open-well format to visualize the cytoskeleton and nuclei, respectively (Figure 7A).
3.4. The flow module does not induce shear activation of neutrophils
During tissue injury or inflammation, leukocytes (e.g., neutrophils) are recruited from the bloodstream to the endothelial surface in response to inflammatory factors and then transmigrate across the barrier in a coordinated process.[29,31,60] It is important to note that neutrophils can become activated upon exposure to shear stress greater than ∼1.5 dynes.cm-2.[31,67,68] Shear activation can change neutrophil responsiveness to inflammatory factors and reduce transmigration efficiency.[53] Thus, to limit shear activation effects within the flow module, we mapped the shear stress distribution along the flow path using our PIV-validated COMSOL model and selected a flow rate where the maximum shear stress was less than the activation threshold of 1.5 dynes.cm-2. For the neutrophil introduction, Q = 10 µL.min−1 was selected to both minimize shear exposure and ensure adequate deposition onto the membrane surface at the initial stage of the transmigration process (Figure S6). Figure 8A showed that the maximum predicted shear stress in the flow path (0.4 dynes.cm−2) occurred at the intersection of vertical access ports and the horizontal channel and was below the shear activation threshold (1.5 dynes.cm−2).
Figure 8.
Shear-induced activation of neutrophils. (A) Maximum shear stress within the flow module occurs at the intersection of vertical access ports and horizontal microchannel (red arrows); based on the simulation, the maximum shear stress is 0.4 dynes.cm−2 at an inlet flow rate of 10 µL.min−1 and neutrophils should not become activated by the fluid dynamics within the flow module. (B) Upon exposure of neutrophils to IL-8 or shear stresses above 1.5 dynes.cm−2, they become activated and shed the surface marker, L-selectin. Quantitative analysis of neutrophil activation based on L-selectin loss shows that the percentage of activated neutrophils in the negative control and flow module are not statistically different, while the positive control with neutrophils treated with 10 ng.ml−1 of the cytokine IL8 shows a significant statistical difference (*p < 0.05, n = 4).
To confirm our model-based prediction that neutrophils introduced at Q=10 μL.min−1 would not exhibit significant shear activation in the flow module, we carried out a flow cytometry study in which the loss of the surface marker, L-selectin, was used as a metric to assess activation (Figure 8B).[51–53] Our results showed no statistical differences between the percentage of activated neutrophils in the population collected from the flow module and the negative control group (Figure 8B, Figure S7), while significant statistical differences were observed compared to the IL8-treated positive control condition (p-value = 0.037, n = 4). Here, Interleukin-8 (IL-8) was used as the positive control because it is a potent activator of neutrophils that results in the shedding of L-selectin and its activation concentration is well-studied in vitro.[69–74] Based on the results, we concluded that our validated COMSOL model could be used to select flow rates that limit shear activation during the introduction of neutrophils in the flow module.
3.5. Neutrophil transmigration dynamics can be visualized under flow conditions
Live monitoring of the neutrophil recruitment and transmigration process can help to provide a deeper understanding of immune cell interactions with the vascular barrier in a physiological setting.[29,31] Using the live imaging and fluid flow capabilities of our platform, we captured the distinct phases of neutrophil transmigration across the endothelial barrier (Video S5). In this assay, the chemoattractant fMLP was introduced to the abluminal channel to promote transmigration.[29,30,49] Upon introduction, the flowing neutrophils came into contact with the stimulated endothelium (Figure 9A). Interactions between the neutrophils and the endothelial monolayer cause the neutrophils to slow (i.e., rolling phase) until their motion is arrested on the endothelium (Figure 9B).[75,76] Neutrophils then crawl on the cell monolayer and can transmigrate across the endothelium (Figure 9C).[75,76] When viewed under transmitted light microscopy, the appearance of the neutrophils changes from “phase bright” (Figure 9D) to “phase dark” (Figure 9E) as they migrate across the monolayer.[29,49] The optical properties of the culture membrane enable the dynamics of transmigration, including partial transmigration, to be visualized in real-time while the addition of fluid flow introduces neutrophils in a physiologically representative environment.
Figure 9.
Transmitted light imaging of neutrophil transmigration dynamics in response to abluminal fMLP stimulation. The HUVEC monolayer was established in the open-well format before the reconfiguration into the fluidic mode. Neutrophils were introduced under flow (left to right) and imaged to show the following steps: (A) rolling along the endothelial cell layer, (B) arrest, (C) crawling on the cell monolayer, and (D, E) transmigrating from the luminal side (pink arrowhead) to the abluminal side of the monolayer (green arrowhead). Scale bars = 10 µm.
Even though the µSiM membrane is thin, it is supported by a rigid silicon component (the blue material in Figure 2B). Because of the small exposed area (0.7 mm × 2 mm) and mechanical stability of the silicon nitride materials, the membrane does not appear to sag or warp.[31,77] We have previously used a dead-ended system to characterize the relationship between inflation pressure and membrane deflection using white light interferometry.[77] In our current system, the maximum normal pressure at the membrane was determined to be 70 Pa using the COMSOL simulation, and a conservative estimate of membrane deflection is < 1µm. This value is consistent with what is shown in Figure 9 and Video S5 where the focal plane does not change and the membrane remains stable over the course of the experiment. Taken as a whole, our platform provides unique opportunities to investigate leukocyte trafficking that is not possible using conventional open-well methods.
3.6. Customization of the m-µSiM components
The mass-produced components of the m-µSiM described in the companion paper have the potential to be customized at the individual laboratory level to enhance the physiological relevance of the barrier tissue model. For example, aligned type-I collagen is of great interest as an extracellular matrix (ECM) material due to its unique microarchitecture and presence in various tissue environments.[78] The incorporation of ECM gels into the abluminal side can enable users to study the post-transmigration guidance of leukocytes in the tissue environment. Our lab has previously shown that local extensional flows can induce long-range collagen fiber alignment.[54] To obtain aligned collagen fibers below the membrane area, we customized the bottom channel of the m-µSiM to include a “segmented” channel geometry and replaced the standard component. As shown in Figure S8, we were able to obtain a 3D collagen gel with aligned fibers below the membrane in the m-µSiM. This simple demonstration highlights how components of m-µSiM can be easily redesigned to introduce new experimental capabilities in different laboratories.
4. Discussion
The goal of this work is to provide an experimental workflow to introduce flow capabilities to the open-well m-µSiM platform. Here, we use a magnetic latching approach that enables reconfiguration between open-well and fluidic modes during the experimental workflow and combines the unique advantages of each approach. In our platform, the barrier is established in the familiar open-well format and reconfigured to a fluidic system during an experiment by adding a flow module. This reconfiguration allows i) the monolayer to experience fluid-induced shear stress that models the physiological environment and ii) supports the introduction of leukocytes to the endothelial monolayer under flow conditions to visualize discrete steps in the recruitment process, including rolling, arrest, crawling, and transmigration. The reconfiguration capability enables users to use established open-well protocols during experiments and also benefit from microfluidic capabilities when desired. Magnetic latching is a unique feature and provides a simple, tool-free assembly approach that allows on-demand switching between open-well and fluidic formats. As a result, different steps of an experiment, including permeability measurements, immunostaining, and RNA extraction can be conducted in the open-well format using established techniques that most laboratories are familiar with.[43]
The reconfiguration capability requires a change in the workflow because the addition of the flow module can damage cells that are seeded uniformly over the membrane and silicon support. Here, we use a cell-seeding stencil that enables controlled positioning of cells directly onto the membrane surface that supports efficient seeding of cells at a known density, which is important because the starting cell concentration influences vascular barrier formation.[42] The tapered well within the stencil increases media capacity (from 10 µl to 30 µl) while limiting bubble trapping that can occur within high aspect ratio, straight-walled reservoirs.[79] Thus, cells can be maintained in long-term culture with the stencil in place if desired. The m-µSiM enables controlled cell seeding in the open-well format before the addition of the flow module and addresses challenges including cell losses along the flow path intrinsic to conventional microfluidic platforms and thus reduces the number of cells used. The stencil also prevents cells from adhering outside of the culture area of the porous membrane. This is particularly important in co-culture models because cells deposited in unwanted locations cannot contribute directly to barrier tissue formation. However, they are extracted during lysis and can confound gene expression studies that aim to probe the influence of cell-cell interactions across the membrane.[80]
Although we used HUVECs in our proof-of-concept experiments, the m-µSiM is compatible with other cell populations including iPSC-derived endothelial cells in co-cultured environments.[43] As a future study, we will incorporate pericytes and astrocytes into our system to mimic the blood-brain barrier for the investigation of sepsis. Furthermore, ECM materials with embedded cells can be introduced to the abluminal side of the platform to mimic the tissue side.
To assess barrier integrity, our companion paper has demonstrated an in situ measurement and sampling based technique to quantify the transport of fluorescently labeled compounds across the barrier[43]. Since our design is reconfigurable, the platform can be converted into the open-well format after flow stimulation and barrier permeability can be assessed using standard methods based on user preference. Our future efforts will establish a transendothelial electrical resistance (TEER) module to provide both electrical impedance and molecular transport measurements across the barrier.
Our companion paper highlights reproducible results using the mass-produced m-µSiM, and here we show reconfiguration capabilities by adding a flow module and customization of the bottom channel of the m-µSiM. Although PDMS was used for the fabrication of the flow module, materials such as moldable polyurethane and thermoplastics can be used if problems related to small-molecule partitioning are identified.[34,81,82] Our design is also compatible with a wide range of materials and does not require the flow module composition to be elastomeric. It is only necessary to have a soft gasket-like material at the interface of the flow module and membrane chip to form a seal. Thus, the main body of the flow module can be fabricated from materials that are very resistant to drug sorption, including polycarbonate (PC),[83] cyclic olefin copolymer (COC),[83–86] Styrene-Ethylene/Butylene-Styrene (SEBS),[87] and tetrafluoroethylene-propylene (FEPM) elastomers.[87]
Our components including, the flow module, stencil, and housings are also amenable to scaled-up fabrication similar to the mass-produced components of the open-well m-µSiM.[43] To enhance the throughput of the platform, we have designed a “functional lid” which provides interior fluidic routing channels that enables us to supply multiple devices simultaneously. As a demonstration, we fabricated a prototype for supplying two devices (Video S6, Figure S9A). This design can be further developed to accommodate 4 and 8 devices. Although the stencil adds an extra step to the workflow, the stencil addition process can be modified by using a rigid carrier that contains an array of stencils (Figure S9B) that can be added to a corresponding array of open wells simultaneously to increase the scalability of the technique.
Given the importance of flows, there are several established techniques to introduce perfusion to microfluidic culture platforms. These are generally categorized into two main groups: i) external pumps (e.g., peristaltic pump, syringe pump) and (ii) integrated pumps (gravity-based pumps,[88–93] electromagnetic pumps,[93–96] pneumatic pumps,[96–100] on-chip peristaltic pump[101]). Our platform is fully compatible with external pumps that are common in most laboratory settings. For example, we used a peristaltic pump for long-term culture and shear-induced alignment of endothelial cells. We then used a syringe pump to introduce neutrophils under continuous flow for the transmigration work. As shown in our previous work,[57] our system is also compatible with a pneumatic system that adjusts the pressure drop across the network and enables rapid flow switching or the introduction of different waveforms.[63] Rigid tubing is commonly used with pneumatic systems to minimize system compliance and additional resistance is added to the fluidic network to allow robust flow control in a low compliance, high resistance network with Q = P.R-1. Programmable syringe or peristaltic pumps can be connected to the device to define the input flow rate using rigid or flexible tubing. Our platform is compatible with a variety of pumping schemes as a result of the O-ring style connector that can be used with rigid and flexible tubing as shown in the functional lid (Figure S9A) and our previous work.[46] This approach allows the system to be easily modified to fit experimental needs of the end user. The functional lid can also be customized at the laboratory level to incorporate electromagnetic, gravity-driven, or other pumps as desired. We anticipate that the experimental flexibility and reconfiguration capabilities presented here will allow more widespread use of flow-enhanced microfluidic barrier models in both bioscience and engineering laboratories.
5. Conclusion
To combine the advantages of open-well and microfluidic devices, we developed a functional flow module that enables on-demand reconfiguration of open-well m-µSiM into a fluidic mode. This reconfigurable platform provides not only conventional open-well cell seeding and barrier establishment but also microfluidic capabilities to introduce controlled flows and secondary cell populations. This unique platform utilizes reversible magnetic latching, allowing the flow module to be removed and the barrier directly accessed for downstream experiments. Users have the flexibility to easily switch between different device modes during an experiment and carry out each experimental step in their preferred mode. This platform includes open-well format cell seeding, fluid flow, live imaging, and direct access to the barrier, whereas all prior platforms lack one or more of these features. Since users can select the device format, perfusion technique, and protocols they are most comfortable with, we anticipate that this device will enable more widespread use of flow-enhanced barrier models in both engineering and bioscience laboratories.
Supplementary Material
Acknowledgments
This research was funded by the National Institute of Health under award numbers R43GM137651, R61HL154249, and UG3TR003281. The authors thank prof. Karin Wuertz-Kozak, Dr. Andrea De Pieri, and Petra Cazzanelli for help with the gene expression studies, Xian Boles from the RIT Medical Illustration MFA program for illustration support, and RIT Machine Shop for aluminum mold fabrication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information
Supporting Information is available in the supplement document.
Conflict of Interest
J.L.M. and T.R.G. are co-founders of SiMPore, Inc. and hold an equity interest in the company. SiMPore is commercializing the ultrathin silicon-based technologies including the membranes used in this study.
References
- [1].Vera D, García-Díaz M, Torras N, Álvarez M, Villa R, Martinez E, ACS Appl Mater Interfaces 2021, 13, 13920. [DOI] [PubMed] [Google Scholar]
- [2].Yu F, Selva Kumar ND, Choudhury D, Foo LC, Ng SH, Drug Discovery Today 2018, 23, 815. [DOI] [PubMed] [Google Scholar]
- [3].Claesson-Welsh L, Dejana E, McDonald DM, Trends in Molecular Medicine 2021, 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rajendran P, Rengarajan T, Thangavel J, Nishigaki Y, Sakthisekaran D, Sethi G, Nishigaki I, Int J Biol Sci 2013, 9, 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu M, Chen G, Fu W, Liao M, Frank JA, Bower KA, Fang S, Zhang Z, Shi X, Luo J, Toxicological Sciences 2012, 127, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chistiakov DA, Orekhov AN, Bobryshev YV, Frontiers in Physiology 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sweeney MD, Kisler K, Montagne A, Toga AW, Zlokovic BV, Nat Neurosci 2018, 21, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kaisar MA, Sajja RK, Prasad S, Abhyankar VV, Liles T, Cucullo L, Expert Opinion on Drug Discovery 2017, 12, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ingber DE, Advanced Science 2020, 7, 2002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Prashanth A, Donaghy H, Stoner SP, Hudson AL, Wheeler HR, Diakos CI, Howell VM, Grau GE, McKelvey KJ, Cancers 2021, 13, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kaisar MA, Abhyankar VV, Cucullo L, in Blood-Brain Barrier (Ed: Barichello T), Springer, New York, NY, 2019, pp. 55–70. [Google Scholar]
- [12].Stone NL, England TJ, O’Sullivan SE, Frontiers in Cellular Neuroscience 2019, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hatherell K, Couraud P-O, Romero IA, Weksler B, Pilkington GJ, Journal of Neuroscience Methods 2011, 199, 223. [DOI] [PubMed] [Google Scholar]
- [14].Kasper J, Hermanns MI, Bantz C, Maskos M, Stauber R, Pohl C, Unger RE, Kirkpatrick JC, Part Fibre Toxicol 2011, 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Janga H, Cassidy L, Wang F, Spengler D, Oestern-Fitschen S, Krause MF, Seekamp A, Tholey A, Fuchs S, Journal of Cellular and Molecular Medicine 2018, 22, 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moriyama T, Sasaki K, Karasawa K, Uchida K, Nitta K, Journal of Cellular Physiology 2017, 232, 3565. [DOI] [PubMed] [Google Scholar]
- [17].Gleeson JP, Estrada HQ, Yamashita M, Svendsen CN, Targan SR, Barrett RJ, International Journal of Molecular Sciences 2020, 21, 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pell TJ, Gray MB, Hopkins SJ, Kasprowicz R, Porter JD, Reeves T, Rowan WC, Singh K, Tvermosegaard KB, Yaqub N, et al. , SLAS DISCOVERY: Advancing the Science of Drug Discovery 2021, 26, 909. [DOI] [PubMed] [Google Scholar]
- [19].Merino A, Sablik M, Korevaar SS, López-Iglesias C, Ortiz-Virumbrales M, Baan CC, Lombardo E, Hoogduijn MJ, Frontiers in Immunology 2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wittchen ES, Worthylake RA, Kelly P, Casey PJ, Quilliam LA, Burridge K, Journal of Biological Chemistry 2005, 280, 11675. [DOI] [PubMed] [Google Scholar]
- [21].Wewer C, Seibt A, Wolburg H, Greune L, Schmidt MA, Berger J, Galla H-J, Quitsch U, Schwerk C, Schroten H, et al. , J Neuroinflammation 2011, 8, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ingber DE, Nat Rev Genet 2022, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Walter FR, Valkai S, Kincses A, Petneházi A, Czeller T, Veszelka S, Ormos P, Deli MA, Dér A, Sensors and Actuators B: Chemical 2016, 222, 1209. [Google Scholar]
- [24].Wang YI, Abaci HE, Shuler ML, Biotechnology and Bioengineering 2017, 114, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang JD, Khafagy E-S, Khanafer K, Takayama S, ElSayed MEH, Mol. Pharmaceutics 2016, 13, 895. [DOI] [PubMed] [Google Scholar]
- [26].Booth R, Kim H, Lab Chip 2012, 12, 1784. [DOI] [PubMed] [Google Scholar]
- [27].Frost TS, Jiang L, Lynch RM, Zohar Y, Micromachines 2019, 10, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chang S-H, Ko P-L, Liao W-H, Peng C-C, Tung Y-C, Micromachines 2021, 12, 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Salminen AT, Zhang J, Madejski GR, Khire TS, Waugh RE, McGrath JL, Gaborski TR, Small 2019, 15, 1804111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Khire TS, Salminen AT, Swamy H, Lucas KS, McCloskey MC, Ajalik RE, Chung HH, Gaborski TR, Waugh RE, Glading AJ, et al. , Cell Mol Bioeng 2020, 13, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mossu A, Rosito M, Khire T, Li Chung H, Nishihara H, Gruber I, Luke E, Dehouck L, Sallusto F, Gosselet F, et al. , J Cereb Blood Flow Metab 2019, 39, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET, Beebe DJ, Lab Chip 2009, 9, 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhou M, Zhang X, Wen X, Wu T, Wang W, Yang M, Wang J, Fang M, Lin B, Lin H, Sci Rep 2016, 6, 31771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ishahak M, Hill J, Amin Q, Wubker L, Hernandez A, Mitrofanova A, Sloan A, Fornoni A, Agarwal A, Frontiers in Bioengineering and Biotechnology 2020, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Loskill P, Sezhian T, Tharp KM, Lee-Montiel FT, Jeeawoody S, Mae Reese W, Zushin P-JH, Stahl A, Healy KE, Lab on a Chip 2017, 17, 1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Vedula EM, Alonso JL, Arnaout MA, Charest JL, PLOS ONE 2017, 12, e0184330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sellgren KL, Hawkins BT, Grego S, Biomicrofluidics 2015, 9, 061102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nalayanda DD, Wang Q, Fulton WB, Wang T-H, Abdullah F, Journal of Pediatric Surgery 2010, 45, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sharma K, Dhar N, Thacker VV, Simonet TM, Signorino-Gelo F, Knott GW, McKinney JD, eLife 2021, 10, e66481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gastfriend BD, Palecek SP, Shusta EV, Current Opinion in Biomedical Engineering 2018, 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wilson HK, Canfield SG, Hjortness MK, Palecek SP, Shusta EV, Fluids and Barriers of the CNS 2015, 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McCloskey MC, Kasap P, Ahmad SD, Su S-H, Chen K, Mansouri M, Ramesh N, Nishihara H, Belyaev Y, Abhyankar VV, et al. , Advanced Healthcare Materials 2022, n/a, 2200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM, ELECTROPHORESIS 2000, 21, 27. [DOI] [PubMed] [Google Scholar]
- [45].Ahmed A, Joshi IM, Larson S, Mansouri M, Gholizadeh S, Allahyari Z, Forouzandeh F, Borkholder DA, Gaborski TR, Abhyankar VV, Advanced Materials Technologies 2021, 6, 2001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Williams MJ, Lee NK, Mylott JA, Mazzola N, Ahmed A, Abhyankar VV, Micromachines 2019, 10, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Abhyankar VV, Wu M, Koh C-Y, Hatch AV, PLOS ONE 2016, 11, e0156341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hasan MR, Peri SSS, Sabane VP, Mansur N, Gao JX, Nguyen KT, Weidanz JA, Iqbal SM, Abhyankar VV, Biomed. Phys. Eng. Express 2018, 4, 025015. [Google Scholar]
- [49].Salminen AT, Tithof J, Izhiman Y, Masters EA, McCloskey MC, Gaborski TR, Kelley DH, Pietropaoli AP, Waugh RE, McGrath JL, Integrative Biology 2020, 12, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jaccard P, New Phytologist 1912, 11, 37. [Google Scholar]
- [51].Ivetic A, Cell Tissue Res 2018, 371, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Von Andrian UH, Hansell P, Chambers JD, Berger EM, Torres Filho I, Butcher EC, Arfors KE, American Journal of Physiology-Heart and Circulatory Physiology 1992, 263, H1034. [DOI] [PubMed] [Google Scholar]
- [53].Mitchell MJ, Lin KS, King MR, Biophysical Journal 2014, 106, 2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ahmed A, Mansouri M, Joshi IM, Byerley AM, Day SW, Gaborski TR, Abhyankar VV, Biofabrication 2022, 14, 035019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zittrell F, CircHist - Circular / Polar / Angle Histogram (Https://Github.Com/Zifredder/CircHist), 2022. [Google Scholar]
- [56].Bredfeldt JS, Liu Y, Conklin MW, Keely PJ, Mackie TR, Eliceiri KW, J Pathol Inform 2014, 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hsu M-C, Mansouri M, Ahamed NNN, Larson SM, Joshi IM, Ahmed A, Borkholder DA, Abhyankar VV, Sci Rep 2022, 12, 10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mai J, Abhyankar VV, Piccini ME, Olano JP, Willson R, Hatch AV, Biosensors and Bioelectronics 2014, 54, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Abhyankar VV, Hatch AV, Advanced Healthcare Materials 2012, 1, 773. [DOI] [PubMed] [Google Scholar]
- [60].Wettschureck N, Strilic B, Offermanns S, Physiological Reviews 2019, 99, 1467. [DOI] [PubMed] [Google Scholar]
- [61].Dewey CF Jr., Bussolari SR, Gimbrone MA Jr., Davies PF, Journal of Biomechanical Engineering 1981, 103, 177. [DOI] [PubMed] [Google Scholar]
- [62].Ashpole NE, Overby DR, Ethier CR, Stamer WD, Investigative Ophthalmology & Visual Science 2014, 55, 8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Recktenwald SM, Wagner C, John T, Lab Chip 2021, 21, 2605. [DOI] [PubMed] [Google Scholar]
- [64].Nayak L, Lin Z, Jain MK, Antioxidants & Redox Signaling 2011, 15, 1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hattori K, Munehira Y, Kobayashi H, Satoh T, Sugiura S, Kanamori T, Journal of Bioscience and Bioengineering 2014, 118, 327. [DOI] [PubMed] [Google Scholar]
- [66].Kong X, Qu X, Li B, Wang Z, Chao Y, Jiang X, Wu W, Chen S-L, Molecular Medicine Reports 2017, 15, 908. [DOI] [PubMed] [Google Scholar]
- [67].Moazzam F, DeLano FA, Zweifach BW, Schmid-Schönbein GW, Proceedings of the National Academy of Sciences 1997, 94, 5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Fukuda S, Schmid-Schönbein GW, Proceedings of the National Academy of Sciences 2003, 100, 13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ball CJ, Reiffel AJ, Chintalapani S, Kim M, Spector JA, King MR, Plastic and Reconstructive Surgery 2013, 131, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Brennan K, Zheng J, in XPharm: The Comprehensive Pharmacology Reference (Eds: Enna SJ, Bylund DB), Elsevier, New York, 2007, pp. 1–4. [Google Scholar]
- [71].Bernhard S, Hug S, Stratmann AEP, Erber M, Vidoni L, Knapp CL, Thomaß BD, Fauler M, Nilsson B, Ekdahl KN, et al. , JIN 2021, 13, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hafezi-Moghadam A, Thomas KL, Prorock AJ, Huo Y, Ley K, Journal of Experimental Medicine 2001, 193, 863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gane J, Stockley R, Thorax 2012, 67, 553. [DOI] [PubMed] [Google Scholar]
- [74].Drost EM, MacNee W, Eur J Immunol 2002, 32, 393. [DOI] [PubMed] [Google Scholar]
- [75].Williams MR, Azcutia V, Newton G, Alcaide P, Luscinskas FW, Trends in Immunology 2011, 32, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Castro Dias M, Odriozola Quesada A, Soldati S, Bösch F, Gruber I, Hildbrand T, Sönmez D, Khire T, Witz G, McGrath JL, et al. , Journal of Cell Science 2021, 134, jcs253880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gillmer SR, Fang DZ, Wayson SE, Winans JD, Abdolrahim N, DesOrmeaux J-PS, Getpreecharsawas J, Ellis JD, Fauchet PM, McGrath JL, Thin Solid Films 2017, 631, 152. [Google Scholar]
- [78].Ahmed A, Joshi IM, Mansouri M, Ahamed NNN, Hsu M-C, Gaborski TR, Abhyankar VV, American Journal of Physiology-Cell Physiology 2021, 320, C1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jensen MJ, Goranovi G, Bruus H, J. Micromech. Microeng. 2004, 14, 876. [Google Scholar]
- [80].Rogers MT, Gard AL, Gaibler R, Mulhern TJ, Strelnikov R, Azizgolshani H, Cain BP, Isenberg BC, Haroutunian NJ, Raustad NE, et al. , Sci Rep 2021, 11, 12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Toepke MW, Beebe DJ, Lab Chip 2006, 6, 1484. [DOI] [PubMed] [Google Scholar]
- [82].Vunjak-Novakovic G, Ronaldson-Bouchard K, Radisic M, Cell 2021, 184, 4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].van Midwoud PM, Janse A, Merema MT, Groothuis GMM, Verpoorte E, Anal. Chem. 2012, 84, 3938. [DOI] [PubMed] [Google Scholar]
- [84].Bale SS, Manoppo A, Thompson R, Markoski A, Coppeta J, Cain B, Haroutunian N, Newlin V, Spencer A, Azizgolshani H, et al. , Biotechnology and Bioengineering 2019, 116, 3409. [DOI] [PubMed] [Google Scholar]
- [85].Jeon JS, Chung S, Kamm RD, Charest JL, Biomed Microdevices 2011, 13, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Moore N, Doty D, Zielstorff M, Kariv I, Moy LY, Gimbel A, Chevillet JR, Lowry N, Santos J, Mott V, et al. , Lab Chip 2018, 18, 1844. [DOI] [PubMed] [Google Scholar]
- [87].Campbell SB, Wu Q, Yazbeck J, Liu C, Okhovatian S, Radisic M, ACS Biomater. Sci. Eng. 2021, 7, 2880. [DOI] [PubMed] [Google Scholar]
- [88].Wang YI, Abaci HE, Shuler ML, Biotechnology and Bioengineering 2017, 114, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Esch MB, Ueno H, Applegate DR, Shuler ML, Lab Chip 2016, 16, 2719. [DOI] [PubMed] [Google Scholar]
- [90].Lee SY, Byun HJ, Choi H, Won J-I, Han J, Park S, Kim D, Sung JH, Biotechnol Bioproc E 2022, 27, 221. [Google Scholar]
- [91].Yang Y, Fathi P, Holland G, Pan D, Wang NS, Esch MB, Lab Chip 2019, 19, 3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Miller PG, Shuler ML, Biotechnology and Bioengineering 2016, 113, 2213. [DOI] [PubMed] [Google Scholar]
- [93].Wang YI, Carmona C, Hickman JJ, Shuler ML, Advanced Healthcare Materials 2018, 7, 1701000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Coppeta JR, Mescher MJ, Isenberg BC, Spencer AJ, Kim ES, Lever AR, Mulhern TJ, Prantil-Baun R, Comolli JC, Borenstein JT, Lab Chip 2016, 17, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tandon V, Kang WS, Robbins TA, Spencer AJ, Kim ES, McKenna MJ, Kujawa SG, Fiering J, Pararas EEL, Mescher MJ, et al. , Lab Chip 2016, 16, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, McKinnon KE, Dokic D, Rashedi AS, Haisenleder DJ, et al. , Nat Commun 2017, 8, 14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Tan K, Keegan P, Rogers M, Lu M, Gosset JR, Charest J, Bale SS, Lab Chip 2019, 19, 1556. [DOI] [PubMed] [Google Scholar]
- [98].Azizgolshani H, Coppeta JR, Vedula EM, Marr EE, Cain BP, Luu RJ, Lech MP, Kann SH, Mulhern TJ, Tandon V, et al. , Lab Chip 2021, 21, 1454. [DOI] [PubMed] [Google Scholar]
- [99].Zhang J, Huang Y-J, Yoon JY, Kemmitt J, Wright C, Schneider K, Sphabmixay P, Hernandez-Gordillo V, Holcomb SJ, Bhushan B, et al. , Med 2021, 2, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, Freake D, Kirschner J, Maass C, Tsamandouras N, et al. , Sci Rep 2018, 8, 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, Lindner M, Drewell C, Bauer S, Thomas A, et al. , Lab Chip 2015, 15, 2688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.