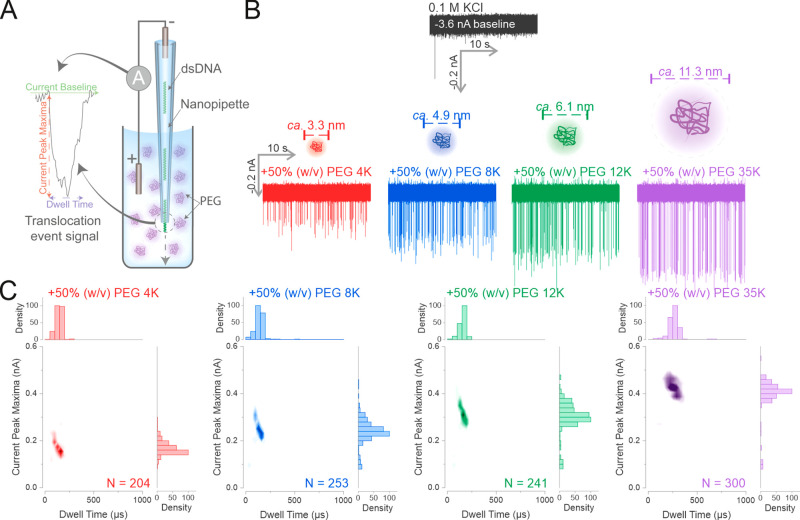
Figure 1.
Increasing the MW of PEG enhances the detection of dsDNA by increasing the SNR. (A) Schematic of the polymer–electrolyte solid-state nanopore setup. The dsDNA is diluted in 0.1 M KCl to 0.3 nM and used to fill the nanopipette. The nanopipette is then immersed into a 50% (w/v) PEG bath containing 0.1 M electrolyte and a negative voltage is applied to drive the translocation of the dsDNA. Each translocation of the dsDNA generates a conductive translocation event signal with two main characteristics: the current peak maxima (the change of current level from the current baseline) and the dwell time (the time it requires for the current to return to baseline). (B) The nanopipette was filled with 0.3 nM of a 4.8 kbp dsDNA diluted in 0.1 M KCl, and immersed into a 0.1 M KCl bath. A voltage of −500 mV was appliedto drive the dsDNA translocations. The same procedure was repeated in 0.1 M KCl baths containing PEGs with different molecular weights as stated. The calculated hydrodynamic diameters for the different molecular weight PEGs are shown.51 (C) The recorded translocation events in the KCl PEG bath of (B) were used to generate density scatter plots and histograms. Four independent nanopore experiments were performed. N is the number of events detected.