Figure 3.
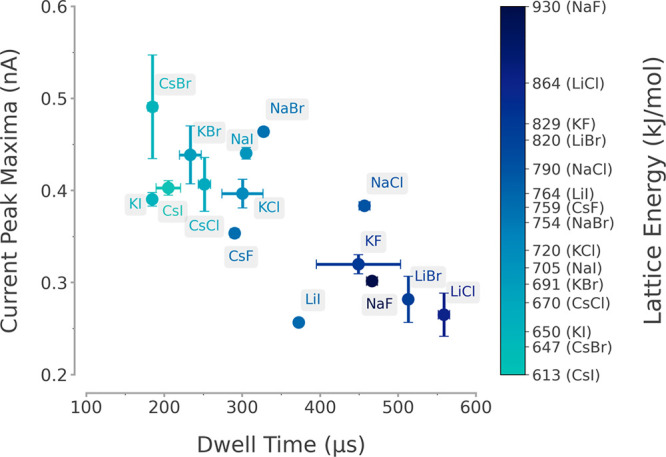
Influence of different alkali metal halide salts on the translocation event signals of 4.8 kb linearized dsDNA. The dsDNA was diluted to 0.3 nM in 0.1 M KCl, and this solution was used to fill the nanopipettes. Different 0.1 M alkali metal halides were used to generate the PEG 35K bath; these were LiCl, LiBr, LiI, NaF, NaCl, NaBr, NaI, KF, KCl, KBr, KI, CsF, CsCl, CsBr, and CsI. The KCl diluted dsDNA nanopipettes were immersed into the different metal halide PEG 35K baths, and −500 mV was applied to drive the dsDNA translocations. The plot displays the mean current peak maxima and the dwell time, and each salt is color coded with its associated lattice energy. Three independent nanopore experiments were performed for each salt condition.
