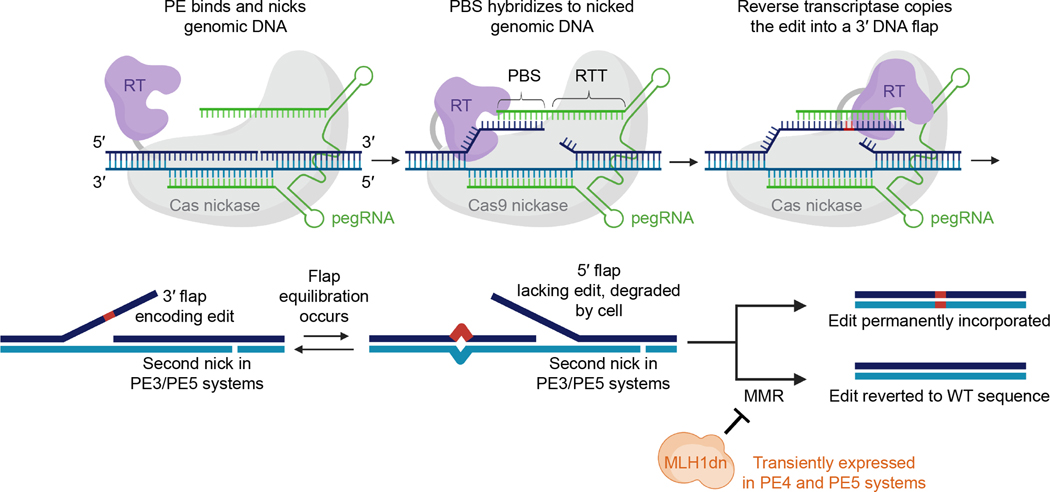
Figure 1. Mechanism of prime editing.
The steps shown above are the putative mechanism for prime editing using various editing systems and an unmodified pegRNA. Cas9 nickase (grey) is recruited to a target DNA site (blue) by a pegRNA (green) and nicks the target site to create a 3′ end of DNA. The primer binding site (PBS) of the pegRNA can then anneal to the genomic DNA flap. This duplex is recognized by a reverse transcriptase (purple), which reverse transcribes nucleotides extending from the target site 3′ end, copying the sequence encoded in the reverse transcription template (RTT) of the pegRNA. Reverse transcription produces a 3′ flap that contains the desired prime edit as well as downstream homology to the rest of the target DNA site. The 3′ flap equilibrates with the corresponding 5′ flap, which does not contain the desired edit. Cellular degradation of the 5′ flap, ligation of the edited 3′ flap into the genome, and repair of the complementary genomic DNA strand by DNA repair or replication results in stable installation of the edit. Prior to repair of the complementary strand, cellular mismatch repair (MMR) can revert the edit back to the unedited sequence. In the PE3 and PE5 systems, a second nick is installed in the complementary strand of DNA, ≥~50 bp away (and typically downstream) from the pegRNA-guided nick. This additional nick biases MMR in favor of editing. In the PE4 and PE5 systems, an engineered dominant-negative MLH1 mutant (MLH1dn, shown in orange) inhibits cellular mismatch repair and thus favors desired prime editing outcomes. This mechanism is based on data collected in previous publications15,30.