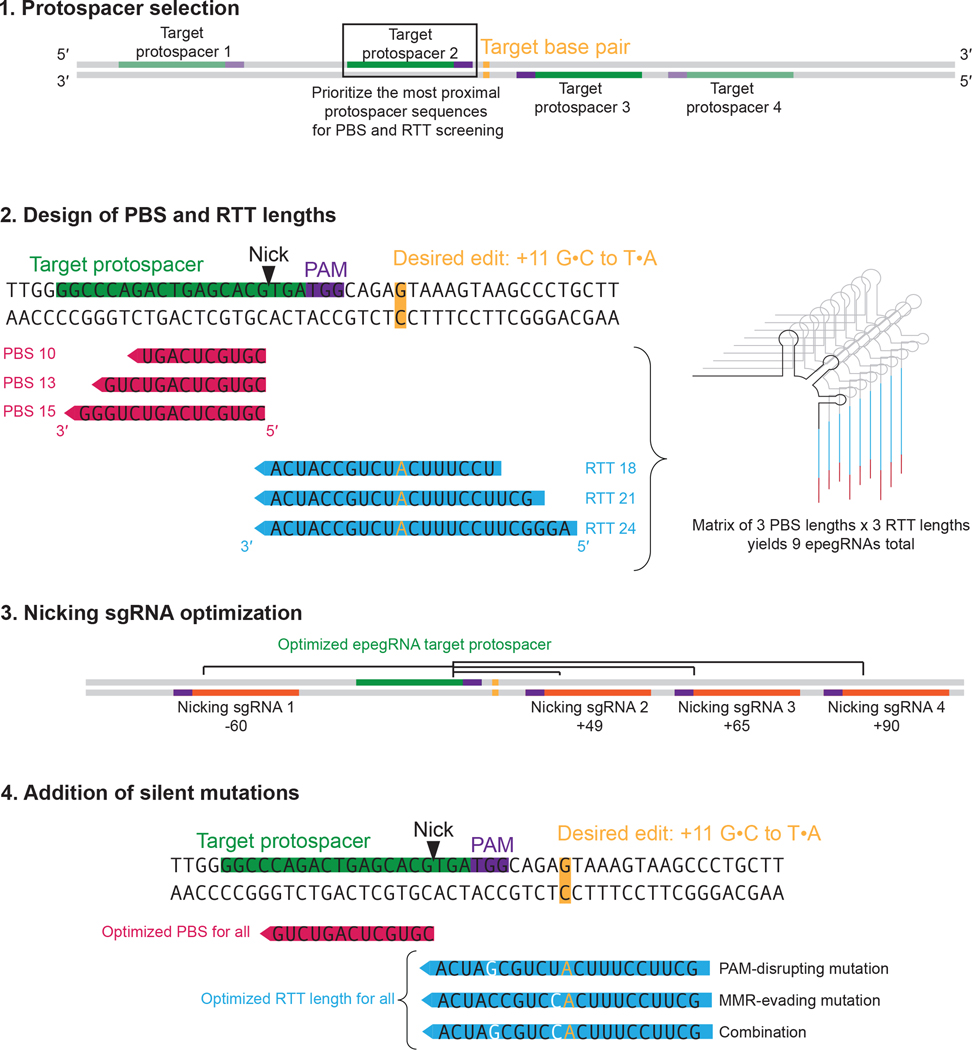
Figure 3. Experimental design of epegRNAs.
Protospacers (shown in green) should first be identified based on available PAM sequences (shown in purple). Of these protospacer candidates, the ones closest to the desired edit (shown in gold) should be tried first. Second, for a minimal initial screen, PBS (shown in pink) lengths of 10, 13, and 15 nt and RTT (shown in blue) lengths that extend at least 7 nt beyond the desired edit are designed. Note: the epegRNA modification is not shown here for simplicity, but it should be included in all pegRNA designs by default. Third, nicking sgRNAs (shown in orange) are designed to target the opposite strand, typically downstream of the initial nick. Finally, PAM-disrupting or silent mutations are identified and added to the RTT of the epegRNAs. This approach combines insights gained from several publications15,30,31.