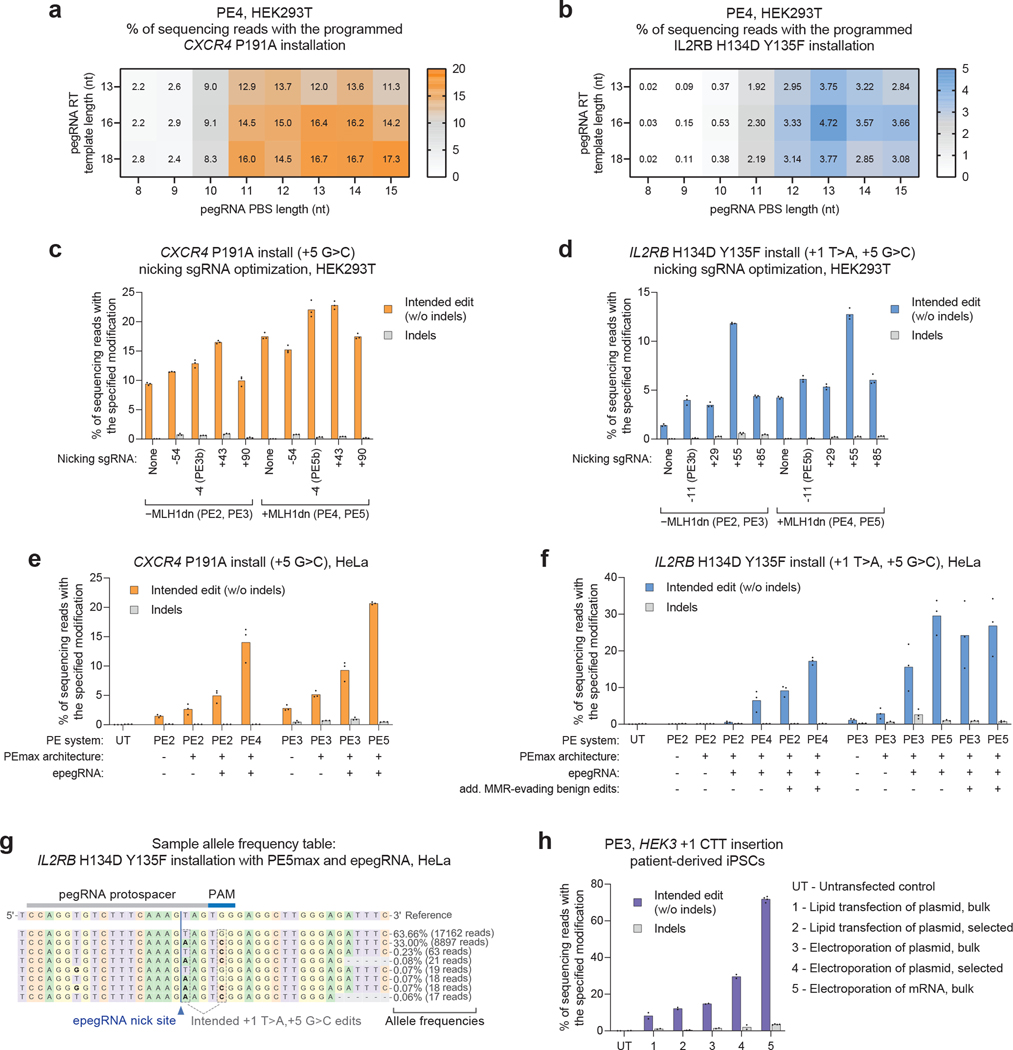
Figure 7. Example results.
(a, b) Heat map showing a PE4 system screen of PBS lengths and RTT lengths for (a) CXCR4 P191A installation and (b) IL2RB H134D + Y135F installation. Note that the optimal PBS and RTT lengths are different between the two sites shown in a and b. Values shown in the heat map cells reflect the mean of n=3 independent replicates. (c, d) Application of nicking sgRNAs at the CXCR4 locus (c) and the IL2RB locus (d). Nicking sgRNAs improve editing in both the PE4 and PE5 system, and MLH1dn improves editing with and without a nicking sgRNA. All values from n=3 independent replicates are shown. (e, f) Editing of the CXCR4 locus (e) and the IL2RB locus (f) in HeLa cells, which are less amenable to prime editing; here, the use of epegRNAs, the PEmax architecture, and MLH1dn dramatically improves editing over the original conditions (PE2 and PE3 with an unmodified pegRNA). All values from n=3 independent replicates are shown. (g) Example allele table generated by CRISPResso2. (h) Example of delivery optimization in patient-derived iPSC cells. Relative to lipid transfection and plasmid electroporation, mRNA electroporation generated a large improvement in editing efficiency. All values from n=3 independent replicates are shown. Data shown in a-h was uniquely collected for this protocol, and is deposited at the NCBI Sequence Read Archive database under PRJNA817825, but experimental techniques are identical to previously reported work15,30,31.