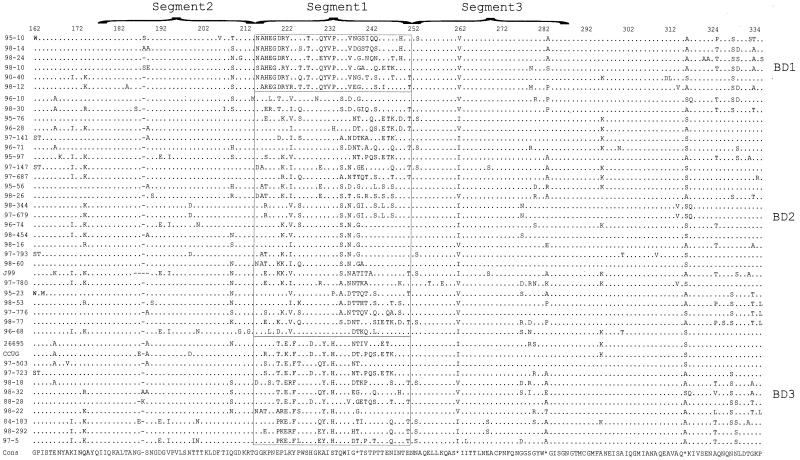
FIG. 5.
Amino acid alignment of babB diversity regions from 42 different H. pylori strains. Amino acid translations of nucleotide sequences were performed using GCG Translate, and sequences were aligned using GCG Pileup. A consensus sequence (based on a plurality of 10) was determined using GCG Pretty, and all the sequences then were displayed using Boxshade 3.2. The dots refer to sites where amino acids match those of the consensus sequence, the hyphens represent deletions, the asterisks represent sites where the consensus sequence is indeterminate, and the boxes are used to separate different allele groups among the babB sequences. Each allele group is based on amino acids 216 to 252 and is represented at the right of each alignment. The babB diversity alleles 1 to 3 are represented as BD1 to BD3, respectively.