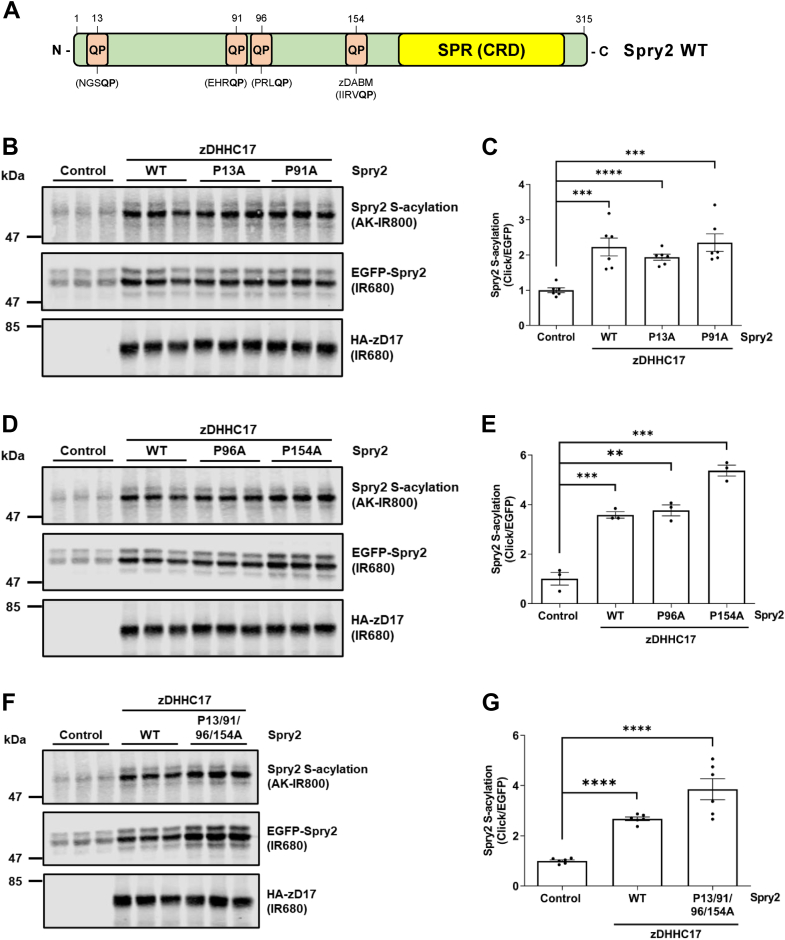
Figure 2.
S-acylation of Spry2 by zDHHC17 does not require zDABM sequences.A, schematic representation of mouse Spry2 protein. All QP dipeptides, which were mutated into “QA,” are shown; the zDABM (IIRVQP) containing proline-154 is indicated. All constructs have EGFP tags appended at the N terminus. SPR denotes the Sprouty domain, which is also referred to as CRD for Cysteine-rich domain. B–G, HEK293T cells were transfected with a plasmid encoding EGFP-tagged Spry2 WT, together with either pEF-BOS-HA (referred to as “control” in the figure) or HA-tagged zDHHC17. Plasmids encoding EGFP-tagged Spry2 P13A, Spry2 P91A, Spry2 P96A, Spry2 P154A, and Spry2 P13/91/96/154A mutants were cotransfected with HA-tagged zDHHC17. Cells were incubated with 100 μM palmitic acid azide for 4 h and labeled proteins reacted with alkyne (AK) IRdye-800 nm. EGFP- and HA-tagged proteins were labeled by immunoblotting using IRdye-680 secondary antibodies. B, D and F, representative images showing Spry2 S-acylation (top; AK-IR800) and Spry2 levels (middle; IR680) detected on the same immunoblot. For zDHHC17, HA (bottom; IR680) was revealed for the same samples on a different immunoblot. The position of the molecular weight markers are shown on the left side of all immunoblots. C, E and G, graphs showing mean Spry2 S-acylation levels after normalization. Error bars represent ± SEM; each replicate is shown with filled circles (n = 3 or 6, different cell samples for each condition). For clarity, only relevant statistical analysis is shown in the figure. Unpaired t test was used to detect significant differences compared to the control sample (∗∗∗∗ denotes p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01). Not shown in the figure: S-acylation of Spry2 P13A, P91A, or P96A versus Spry2 WT was not significant (p > 0.05); S-acylation of both Spry2 mutants containing the P154A substitution was significantly different from Spry2 WT (P154A was p < 0.01 and P13/91/96/154A was p < 0.05).