Figure 3.
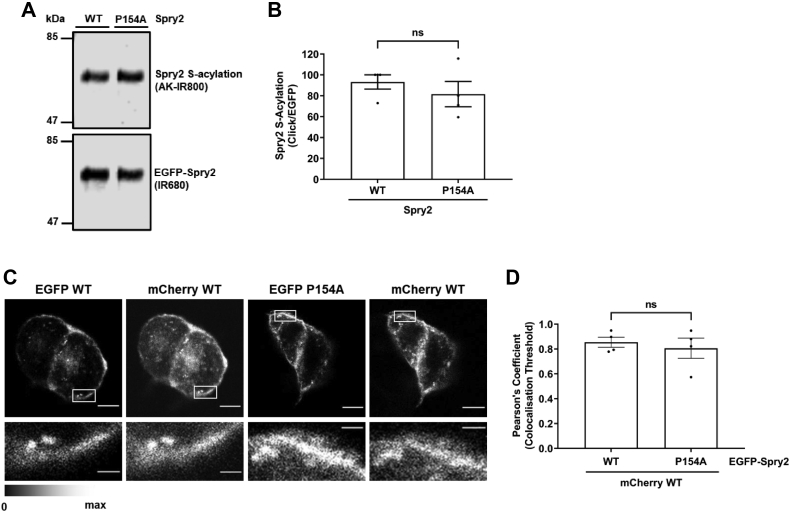
Mutation of P154 in Spry2 does not affect S-acylation or localization in PC12 cells.A, PC12 cells were transfected with either EGFP-Spry2 WT or Spry2 P154A. Cells were incubated with 100 μM palmitic acid azide for 4 h. After cell lysis, labeled proteins were incubated with agarose beads conjugated to an EGFP antibody and later reacted with alkyne (AK) IRdye-800 nm. Representative images showing Spry2 S-acylation (top panel; IR800) and Spry2 expression levels (bottom panel; IR680) detected on the same immunoblot. The position of the molecular weight markers are shown on the left side of the immunoblots. B, graph showing mean Spry2 S-acylation levels after normalization. Error bars represent ± SEM; filled circles represent independent experiments (n = 4, from three independent experiments). An unpaired t test was used to compare S-acylation of Spry2 WT and the P154A mutant (ns denotes nonsignificance i.e. p > 0.05). C, confocal imaging of PC12 cells cotransfected with plasmids encoding EGFP-Spry2 WT or P154A mutant, together with mCherry-Spry2 WT. Representative images for mCherry and EGFP proteins are shown in the figure (upper panels) as well as magnified images of the indicated areas for both channels (bottom panels). The scale bars represent 5 μm for upper full images and 1 μm for the lower magnified images. D, graph showing Pearson’s correlation coefficient (Rtot). Each bar shows mean values of Rtot ± SEM; filled circles represent individual images. Results were analyzed by unpaired t test (ns denotes nonsignificance i.e. p > 0.05, n = 4).