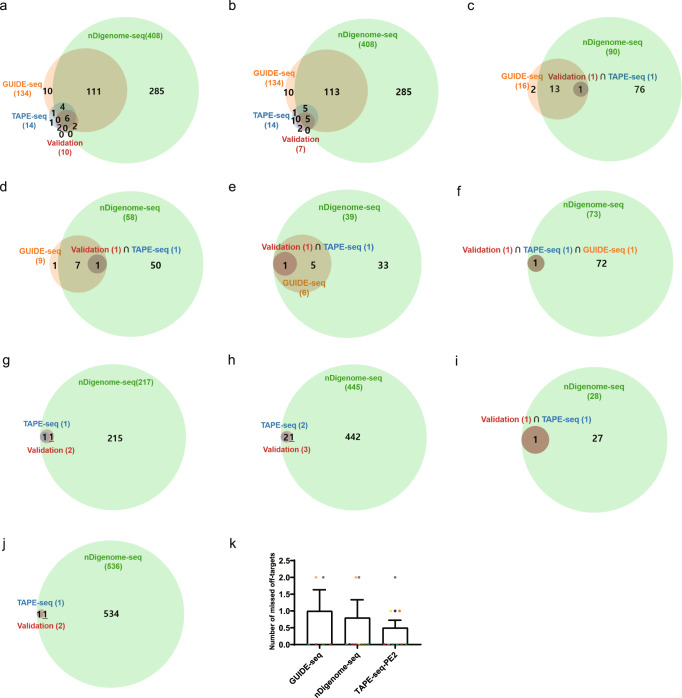
Fig. 2. Comparisons of TAPE-seq prediction results with those from GUIDE-seq and nDigenome-seq.
a–j Venn diagrams showing the number of and overlap between off-target loci predicted by nDigenome-seq, GUIDE-seq, and TAPE-seq and validated off-target sites for a HEK4 (+2 G to T), b HEK4 (+3 TAA ins), c EMX1 (+5 G to T), d FANCF (+6 G to C), e HEK3 (+1 CTT ins), f RNF2 (+6 G to A), g DNMT1 (+6 G to C), h HBB (+4 A to T), i RUNX1 (+6 G to C), and j VEGFA (+5 G to T) pegRNAs. k The number of off-target sites missed by nDigenome-seq (n = 10 independent experiments each represented by different color), GUIDE-seq (n = 6 independent experiments each represented by different color), and TAPE-seq (n = 10 independent experiments each represented by different color). Some numbers have been underlined to differentiate neighboring numbers (g, h, j). The bars represent the mean. Error bars indicate standard deviation (k).