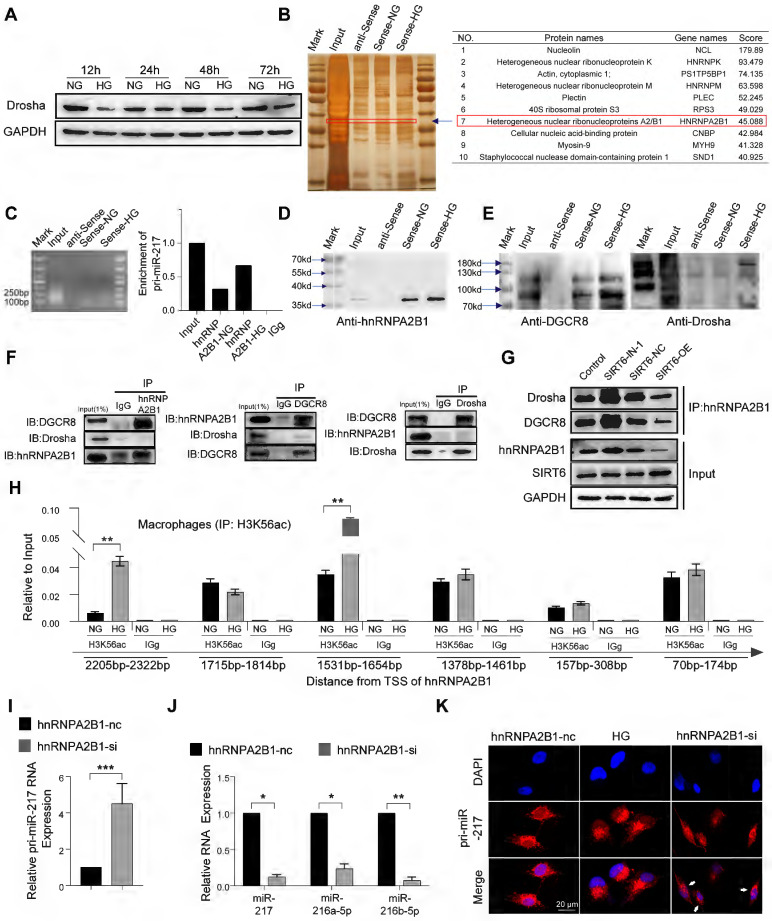
Figure 6.
SIRT6 restrains miR-216a-5p-216b-5p-217 cluster maturation through ''non-canonical'' microprocessor complex. (A) High glucose stimulation inhibits the expression of the Drosha protein in macrophages. (B) Silver staining of pri-miR-217 pulldown in macrophages. List of the top ten differentially expressed proteins identified by mass spectrometry, FDR < 0.05. (C) Expression levels of pri-miR-217 were detected by qRT-PCR after RIP for hnRNPA2B1 in macrophages. (D) Western blotting showed pri-miR-217 pulldown of the hnRNPA2B1. (E) Western blotting showed pri-miR-217 pulldown of the DCGR8 and Drosha. (F) Immunoprecipitation of DGCR8 and Drosha and hnRNPA2B1. (G) Immunoprecipitation of Drosha and DGCR8 using an anti-hnRNPA2B1 antibody. Inhibition of SIRT6 reduces the formation of a ''non-canonical'' microprocessor complex. By contrast, Overexpression of SIRT6 promotes the formation of a ''non-canonical'' microprocessor complex. (H) H3K56ac was enriched around the TSS of hnRNPA2B1 in macrophages by ChIP-qPCR analysis. (I-J) The expression of pri-miR-217 and miR-216a-5p/ miR-216a-5p/ miR-217 in macrophages after si-hnRNPA2B1. (K) FISH localization of pri-miR-217 in macrophages after si-hnRNPA2B1 or high glucose stimulation. U6 and 18S rRNA were used as positive controls for the nuclear and cytoplasmic fractions, respectively. The results were presented as means ± S.D. *p < 0.05; **p < 0.01; ***p > 0.001 by 2-tailed, unpaired Student's t test.