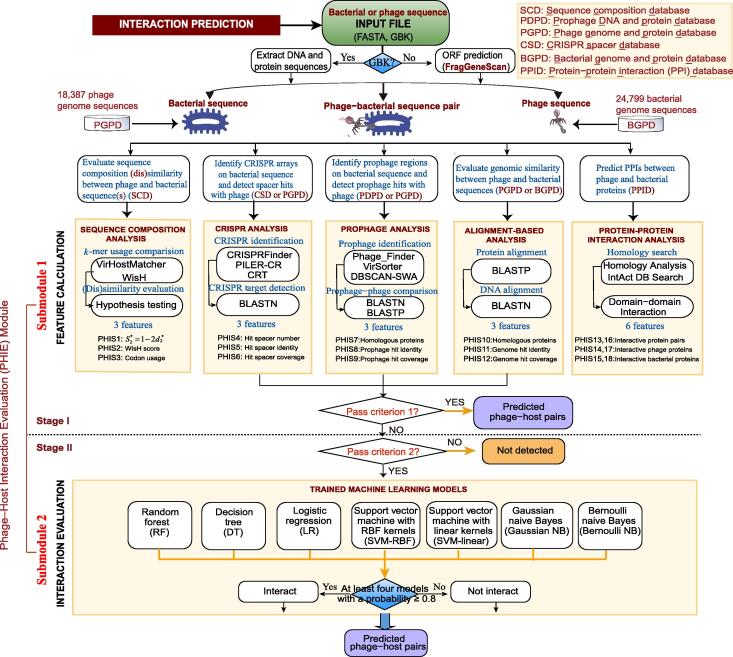
Figure 1.
PHISDetector pipeline for prediction and evaluation of microbe–phage interactions
For candidate phage–bacterial sequence pairs, eighteen PHIS features belonging to five categories are calculated using sequence composition similarity, CRISPR targeting, prophage, genetic homology, and PPI/DDI. Then, a two-stage procedure is performed to predict and evaluate their interactions. In stage I, phage–host pairs with high reliability are detected using criterion 1 and returned directly as final predicted results. In stage II, phage–host pairs with potential PHISs based on criterion 2 are retained and further evaluated using seven well-trained machine learning models including RF, DT, LR, SVM-RBF, SVM-linear, Gaussian NB, and Bernoulli NB, and the phage–host pairs distinguished by at least four models with a probability ≥ 0.8 were returned. PPI, protein–protein interaction; DDI, domain–domain interaction; RF, random forest; DT, decision tree; LR, logistic regression; SVM, support vector machine; RBF, radial basis function; NB, naive Bayes; GBK, GenBank; PHIS, phage–host interaction signal; CRISPR, clustered regularly interspaced short palindromic repeats; ORF, open reading frame; BLASTN, Nucleotide Basic Local Alignment Search Tool; BLASTP, Protein Basic Local Alignment Search Tool; CSD, CRISPR spacer database; PDPD, prophage DNA and protein database; PGPD, phage genome and protein database; SCD, sequence composition database; BGPD, bacterial genome and protein database; PPID, protein–protein interaction database.