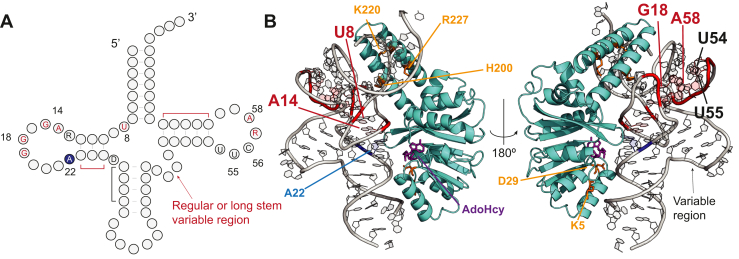
Figure 8.
Structure modeling with the experimental restraints provides a reasonable structure of the TrmK–tRNA complex.A, the tRNA recognition mechanism of TrmK. The nucleotides and stem structure (lines) shown in the secondary structure are recognized by TrmK. The red colored are the critical regions of the tRNA recognition mechanism identified in this study using tRNA-MaP. The nucleotide symbols R and D are purine (G or A) and the nucleotides (G or A or U), respectively. B, the structure of the TrmK–tRNALeu complex was modeled by HDOCK with the input of experimental restraints (see the Experimental procedures section) (63). Amino acid residues of TrmK key for methylation are highlighted in yellow. The color code is the same with (A). The TrmK structure (PDB: 6Q56) and tRNALeu structure (PDB: 2BYT) were used. The Mycoplasma capricolum TrmK structure (transparent) complexed with AdoHcy (purple) was superposed onto the model. MaP, Mutational Profiling.