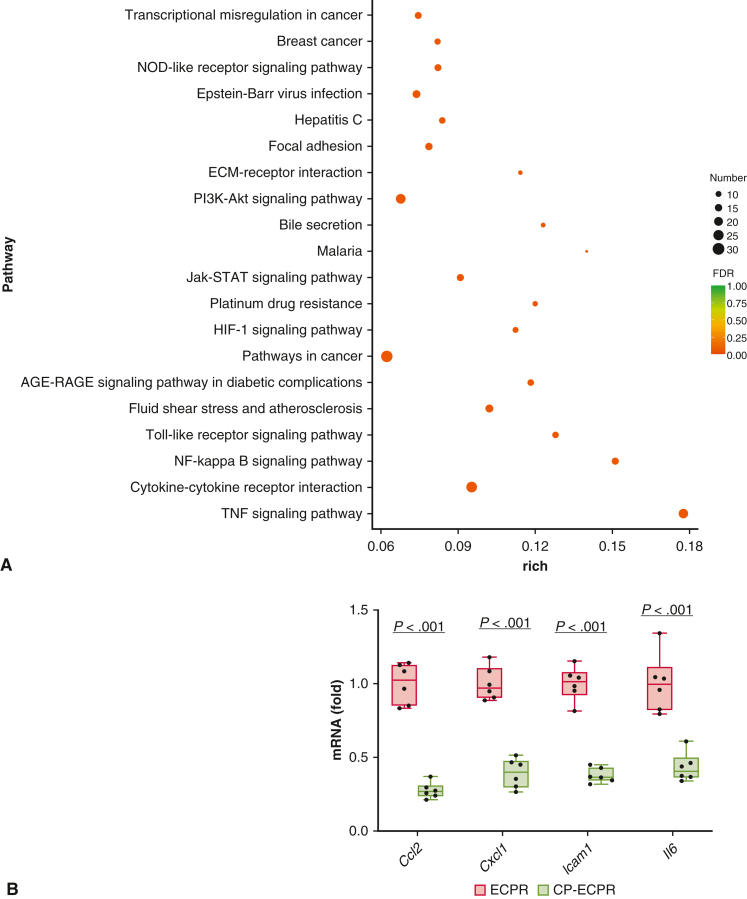
Figure 5.
RNA-sequencing analysis on brain tissues. A, Primary enriched KEGG pathways between ECPR and CP-ECPR groups (n = 3 per group). The y-axis represents the different KEGG categories, and the x-axis represents the rich factor. The size of the dot indicates the number of DEGs involved in the pathway. The color bar represents the P value of the KEGG pathway. B, Transcriptomics data validated by qRT-PCR. Compared with the ECPR group, the mRNA levels of Il6, Ccl2, Cxcl1, and Icam1 in the CP-ECPR group were significantly decreased (n = 6 per group). Graphs represent means ± standard deviation. The box-and-whiskers dot plots includes the minimum value, the lower quartile (25th percentile), the median, the upper quartile (75th percentile), and the maximum value. NOD, Nucleotide oligomerization domain; ECMO, extracorporeal membrane oxygenation; Jak-STAT, Janus tyrosine kinase - signal transducer and activator of transcription; HIF, hypoxia-inducible factor; AGE-RAGE, advanced glycation end products - receptor for advanced glycation end products; NF, nuclear factor; TNF, tumor necrosis factor-α; ECPR, extracorporeal cardiopulmonary resuscitation; CP-ECPR, selective hypothermic cerebral perfusion combined ECPR; DEGs, differentially expressed genes; KEGG, kyoto Encyclopedia of Genes and Genomes; qRT-PCR, quantitative real-time polymerase chain reaction.