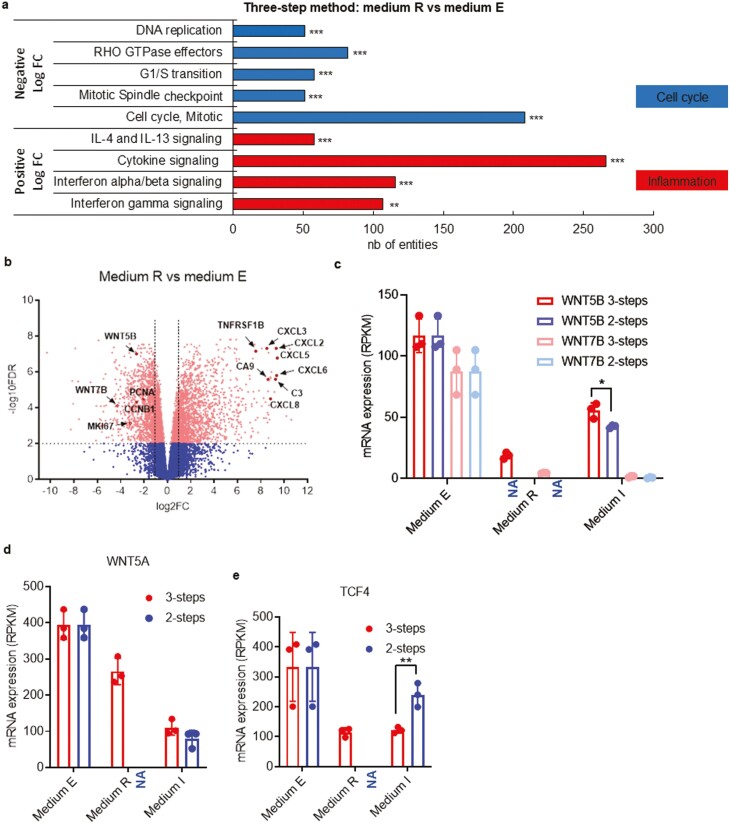
Figure 3.
Inhibition of WNT signalling enhances the production of hyaline matrix components. (a) Gene set enrichment analysis and functional annotation clustering are based on the differential expression levels of MR versus ME in the 3-step method. Histograms show the number of genes for each family of genes for which a significant enrichment was found (number of genes differentially expressed). Top gene sets had lower expression in MR than in ME (negative FC), while bottom gene sets had higher expression in MR (positive FC). (b) Visualization of RNAseq results from chondrocytes in MR versus ME in the 3-step method. The Volcano plot shows results with statistical significance (FDR < .01) versus magnitude of change (>2-fold change). The plot highlights genes that are significantly (scatter above the dotted line) upregulated (right-hand side) and downregulated (left-hand side). In this plot, WNT5B, WNT7B, and MKI67 are decreased in MR versus ME in the 3-step method. Data from RNAseq analysis comparing mRNA expression levels (in RPKM) for WNT5B and WNT7B (c), WNT5A (d), TCF4 (e) genes in the 3-step and 2-step method. RNAseq analysis was made from 3 donor samples. One biological sample per donor was used. In all the study, chondrocytes are used between passage 3 to 7. Of note, the starting material corresponding to the expansion step (step 1) in ME is identical for the 2-step (right-hand side bars) and 3-step (left-hand side bars) method. NA (Not Applicable) refers to the lack of this step in the 2-step method, thus resulting in an absence of data. *P < .05, **P < .01; ***P < .001.