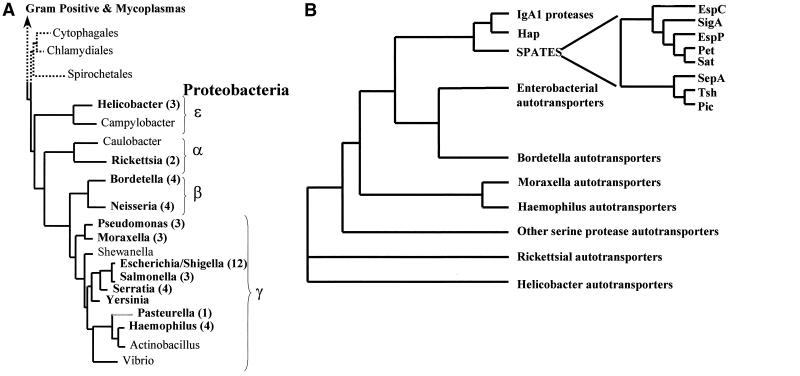
FIG. 2.
Phylogenetic distribution of the autotransporters. (A) Phylogenetic tree of the eubacteria based on 16S rRNA sequences. The tree contains a detailed list of the gram-negative proteobacteria and their subdivisons (identified by Greek symbols) and was adapted from information appearing elsewhere (137) and at the Bergey's Manual Trust website (http://www.cme.msu.edu/Bergeys/btcomments/bt9.pdf). Genera for which autotransporters have been identified and characterized are indicated by boldface. The number of autotransporters identified per genus is given in parentheses after the genus name. (B) Phylogenetic tree of the autotransporter proteins inferred from comparison of the complete amino acid sequences of the polyproteins. The sequences were aligned using the multisequence alignment program CLUSTALX. Phylogenetic relationships and evolutionary distances were calculated, and a dendrogram was constructed using the neighbor-joining method. The positions of the autotransporter proteins within the tree are indicated by families such that the IgA1 proteases from Neisseria meningitidis and N. gonorrhoeae and H. influenzae cluster in the IgA1 protease family, etc.