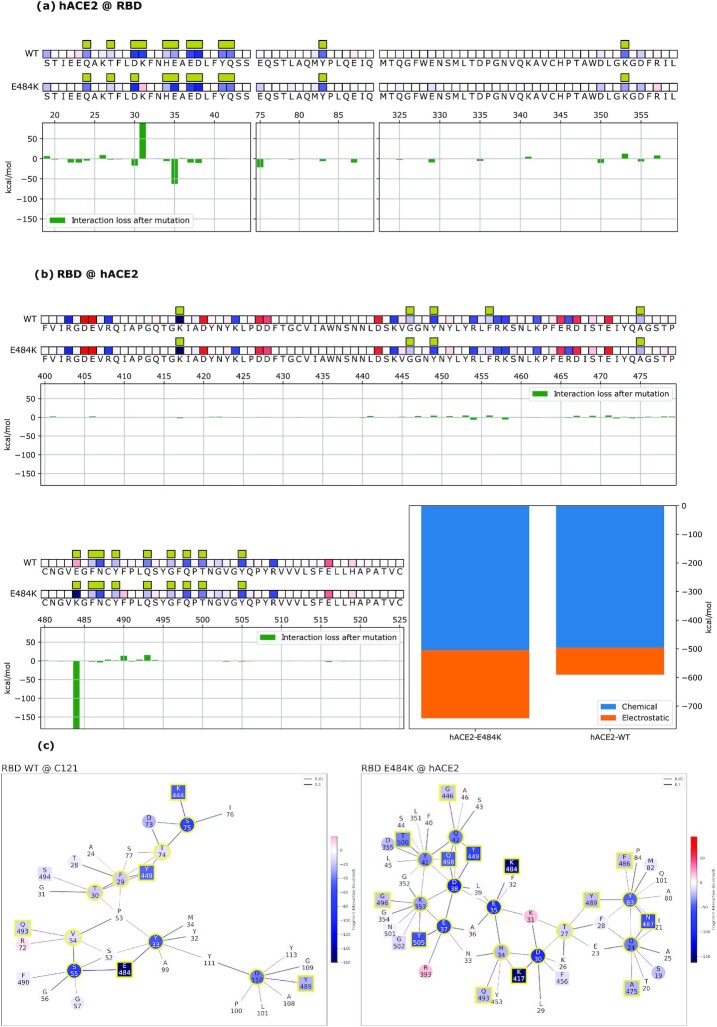
Fig. 4.
Mechanistic characterization of C121 binding to the Wuhan strain spike protein and energetic changes as a result of the E484K spike mutation. Data are plotted on the spike primary structure (a) and on C121’s Heavy-Chain (b) considering the different bindings via the Wuhan spike (WT) and the mutated one (E484K). Amino acids are represented by letters and numbered on the histogram's horizontal axis. Histograms underneath the sequences represent the relative change in binding energy of the second row relative to the first one (Wuhan strain). The bottom right histograms represent the overall binding energy of C121 with the Wuhan spike (left) and the mutated one (right) and its characterization as chemical or electrostatic. The row above each sequence shows the chemical or electrostatic forces as attractive (blue) or repulsive (red), with darker colors indicating stronger effects. Interaction networks with C121 nAbs are shown (c). Network nodes are represented in red (repulsive) or blue (attractive) based on their effect on their counterparts. Residues at the binding interface are highlighted by a yellow outline. Bonds are plotted as purple when intermolecular or black when intramolecular and their thickness is related to the strength of the FBO between residues.