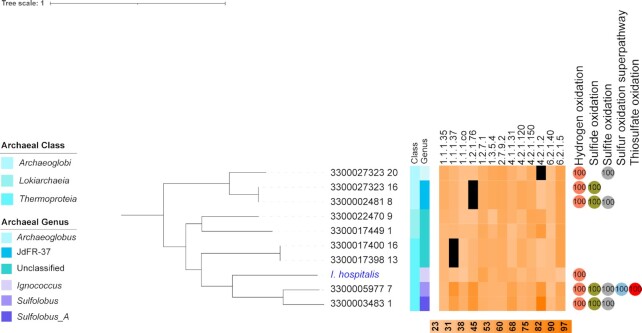
Fig. 5.
Phylogenetic tree showing the distribution of the DC/HB cycle in novel archaeal taxa and the reference genome of I. hospitalis. Identity of the enzymes involved in the pathway against the UniRef database are shown in orange heat scale. 1.1.1.35: S-3-hydroxybutyryl-CoA dehydrogenase; 1.1.1.37: malate dehydrogenase; 1.1.1.co: succinate semialdehyde reductase; 1.2.1.76: succinyl-CoA reductase; 1.2.7.1: pyruvate synthase (ferredoxin-dependent); 1.3.5.4: fumarate reductase; 2.7.9.2: phosphoenolpyruvate synthetase; 4.1.1.31: phosphoenolpyruvate carboxylase; 4.2.1.120: 4-hydroxybutyryl-CoA dehydratase; 4.2.1.150: S,-3-hydroxybutanoyl-CoA dehydrogenase; 4.2.1.2: fumarase; 6.2.1.40: 4-hydroxybutyryl-CoA synthetase; 6.2.1.5: succinyl-CoA synthetase. Numbers in circles refer to completeness of diagnostic pathways for energy generation. Black squares in the heatmap refers to missing enzymes. The tree was rooted with a MAG from the Methanobacteriota, which is not shown here.