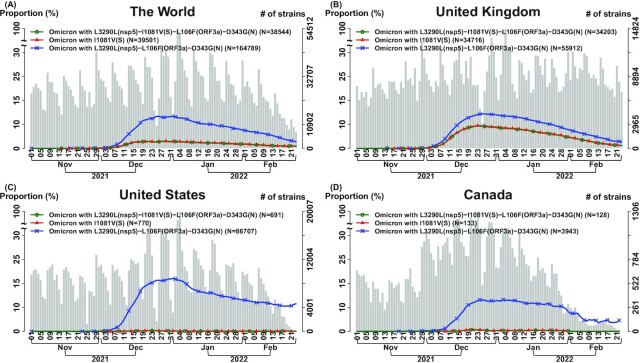
Fig. 5.
Single spike suppressor SNV and set of transmission suppressor SNVs contributes to transmission suppression in Omicron-07 (n = 8,475 K genomes as of 2022 February 23). We analyzed transmission suppression contributed by a single suppressor SNV and a set of suppressor SNVs. Omicron-07 (Fig. 3 and Table S3) is illustrated as an example. In each subfigure, three curves indicate the temporal trajectories for the following three subgroups: (i) “Omicron-07” carries a full set of four transmission suppressor SNVs: L3290L(nsp5)–I1081V(S)–L106F(ORF3a)–D343G(N) (green color and circle symbol); (ii) “Omicron-07 with L3290L(nsp5)–L106F(ORF3a)–D343G(N)” indicates the Omicron strains that they carry the signature SNVs similar to Omicron-07 but miss a transmission suppressor SNV I1081V in the spike protein, i.e. it only carries the suppressor triplet L3290L(nsp5)–L106F(ORF3a)–D343G(N) (blue color and x symbol); (iii) “Omicron with I1081V(S)” indicates the Omicron strains carrying a transmission suppressor SNV in the spike protein I1081V (red color and triangle symbol). The number of total strains per date (gray bar) is displayed with the histogram in the background. “Omicron-07” and “Omicron with I1081V(S)” have very close temporal trajectories, representing that the suppressor SNV I1081V(S) and triplet L3290L(nsp5)–L106F(ORF3a)–D343G(N) co-appeared in the Omicron variants. They have a lower temporal trajectory compared to the one in “Omicron-07 with L3290L(nsp5)–L106F(ORF3a)–D343G(N),” indicating that I1081V(S) and/or L3290L(nsp5)–I1081V(S)–L106F(ORF3a)–D343G(N) have an effect in suppressing a viral transmission of Omicron. More data are needed to distinguish that the suppressing effect is contributed by I1081V solely and/or the full set of transmission suppressor SNVs: L3290L(nsp5)–I1081V(S)–L106F(ORF3a)–D343G(N). (A) The World; (B) The United Kingdom; (C) The United States; (D) Canada.