Figure S3.
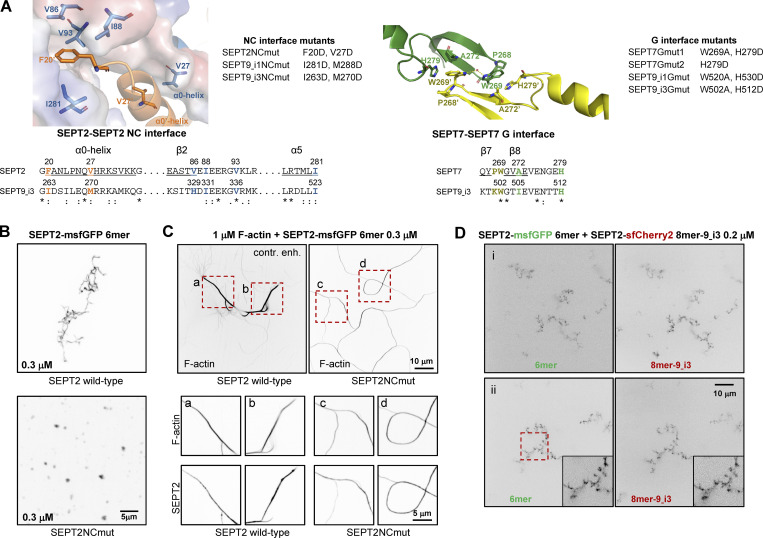
Septin interface mutants used in this study and cell-free reconstitution of septin assembly. (A) Left, top: Conserved residues in the SEPT2-SEPT2 NC interface are shown in the crystal structure of human SEPT2 homodimers (PDB accession no. 2QA5; Sirajuddin et al., 2007). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues F20 from the hook-loop of one SEPT2 subunit (orange) interact with the hydrophobic cleft formed by V86, I88, V93, and I281 of the adjacent SEPT2 subunit (blue). The importance of this phenylalanine in anchoring the α0 helix at the NC interface was emphasized only recently (Cavini et al., 2021). The blue subunit's surface representation highlights the complementary of shape between the two SEPT2 subunits in this interface. The interaction between the α0 helices of each subunit is also stabilized through a hydrophobic interaction between their respective V27. Left, bottom: Sequence alignment of the regions including the residues shown in the NC interface structure for SEPT2 and SEPT9_i3. The structural elements (α0, β2, α5) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties; period, conservation between residues with weakly similar physicochemical properties). We note that the residues described above are strictly or physicochemically conserved (except for V86), highlighting their importance in stabilizing the SEPT2-SEPT2 NC interface. The NC interface mutants used in this study are listed on the right of the crystal structure. A mutation of F20D/I263D is expected to destabilize the hydrophobic pocket depicted above, whereas a V27D/M270D is expected to destabilize the α0 helices interface. Importantly, a strictly conserved aspartate (SEPT2 E90, corresponding to SEPT6 E90 and SEPT7 E102 which are well defined in the cryo-EM structure of the SEPT6-SEPT7 NC interface [Mendonca et al., 2019]) in the loop connecting β2 and β3 is pointing to the hydrophobic cleft where the phenylalanine resides. The F20D mutation is thus expected to result in a repulsion between the aspartate and glutamate and contribute further to the destabilization of the NC interface. Right, top: Conserved residues in the SEPT7-SEPT7 G interface are shown in the crystal structure of human SEPT7 homodimers (PDB accession no. 6N0B; Brognara et al., 2019). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues W269 of one SEPT7 subunit (yellow) interact with residues W269, A272 and H279 in the adjacent SEPT7 subunit (green; Sirajuddin et al., 2007; Zent et al., 2011). W269 from adjacent subunits interact through a water molecule bridge through hydrogen bonds. In addition, each W269 is engaged in π-π interactions with H279 and CH-π interactions with A272 of the opposite subunit. Right, bottom: Sequence alignment of the regions including the residues shown in the G interface structure for SEPT7 and SEPT9_i3. The structural elements (β7, β8) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties). Notice that W269 and H279 are both strictly conserved, showing their importance in stabilizing this interface. The G interface mutants that were used in this study are listed on the right of the crystal structure. The presence of both mutations W269A and H279D in SEPT7 and SEPT9 is expected to destabilize the SEPT7-SEPT7 and SEPT7-SEPT9 G-interfaces. The loss of the aromatic cycle properties in the mutant W269A does not allow the abovementioned critical interactions mediated by the wild-type Trp. W269A is expected to destabilize H279 and potentially change its orientation. In addition, the much smaller size of the alanine will poorly mimic the hydrophobic interaction between W269 and H279, weakening the G-interface. Note that W269 is in the vicinity of Y267 of the same subunit. This tyrosine interacts with the nucleotide buried within the G-interface. Consequently, any mutations destabilizing W269 could dramatically destabilize the overall G-interface because of a domino effect. Similarly, H279D is expected to preclude hydrophobic interactions with W269 and thus destabilize the latter. The single mutation H279D in SEPT7 is expected to destabilize the SEPT7-SEPT7 G-interface when present in both SEPT7 subunits, but not the SEPT7-SEPT9 interface with wild-type SEPT9. (B) Representative spinning disk fluorescence images of septin filament assembly upon polymerization of hexamers in solution at the indicated final protomer concentration. Protomers contained either wild-type SEPT2 (top panel) or SEPT2NCmut (bottom panel). Images use an inverted grayscale. Related to Fig. 2 C. (C) Representative spinning disk fluorescence images of reconstituted actin filaments, polymerizing in the presence of septin hexamers in solution. Protomers contained either wild-type SEPT2 or SEPT2NCmut. Actin filaments are visualized with AlexaFluor568-conjugated phalloidin, and septins with SEPT2-msfGFP. One example of large fields of view are shown for each condition, depicting cross-linking of actin filaments; only actin labeling is shown. The image for actin in the presence of wild-type hexamers is contrast-enhanced on purpose in order to saturate the actin bundles so that weaker-intensity single actin filaments are also visible. Insets on the bottom show higher magnifications of selected regions of interest on the top (dashed squares in red). Two regions of interest (a and b for wild-type SEPT2 and c and d for SEPT2NCmut) are shown in each case, depicting both the actin and septin signals. Scale bars in all large fields of views, 10 μm. Scale bars in all insets, 5 μm. (D) Representative spinning disk fluorescence images of septin filament assembly upon co-polymerization of hexamers containing SEPT2-msfGFP and octamers-9_i3 containing SEPT2-sfCherry2 at the indicated final protomer concentration. Images use an inverted grayscale. Scale bars in all large fields of views, 10 μm.