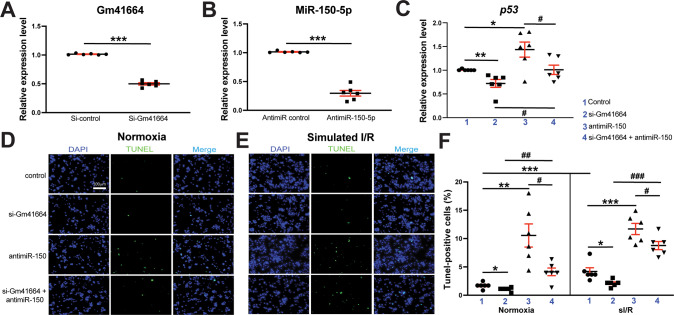
Fig. 7. MiR-150 is required for Gm41664-dependent induction of cardiomyocyte apoptosis.
A, B HL-1 cells were transfected with control scramble siRNA (si-control) or Gm41664 siRNA (si-Gm41664) (A) and with anti-miR control scramble or anti-miR-150 (B). QRT-PCR for Gm41664 (A) or miR-150 (B) was conducted to check the knockdown efficiency. Data were normalized to Gapdh (A) or U6 snRNA (B) and expressed relative to controls. N = 6 per group. Unpaired two-tailed t-test. ***P < 0.001 vs. si-control or anti-miR control. C QRT-PCR expression analysis of pro-apoptotic p53 in cardiomyocytes (CMs) transfected with 4 different groups as indicated. N = 6. p53 expression compared to Gapdh was calculated using 2−ΔΔCt, and data are presented as fold induction of p53 expression levels normalized to control (si-control or anti-miR control). One-way ANOVA with Tukey multiple comparison test. *P < 0.05 or **P < 0.01 vs. control. #P < 0.05 vs. si-Gm41664 + anti-miR-150. D–F MiR-150 knockdown reverses the anti-apoptotic effects of si-Gm41664 in CMs. CMs were transfected as indicated and subjected to in vitro simulation of I/R (hypoxia/reoxygenation) [sI/R (H/R)]. TUNEL assays were then conducted in both normoxic (D, F) and sI/R conditions (E, F). The percentage of apoptotic nuclei (green) was calculated after the normalization of total nuclei (blue). Scale bar = 100 μm. N = 6. One-way ANOVA with Tukey multiple comparison test. *P < 0.05, **P < 0.01, or ***P < 0.001 vs. control. #P < 0.05, ##P < 0.01, or ###P < 0.001 vs. si-Gm41664 + anti-miR-150. All data are presented as mean ± SEM.