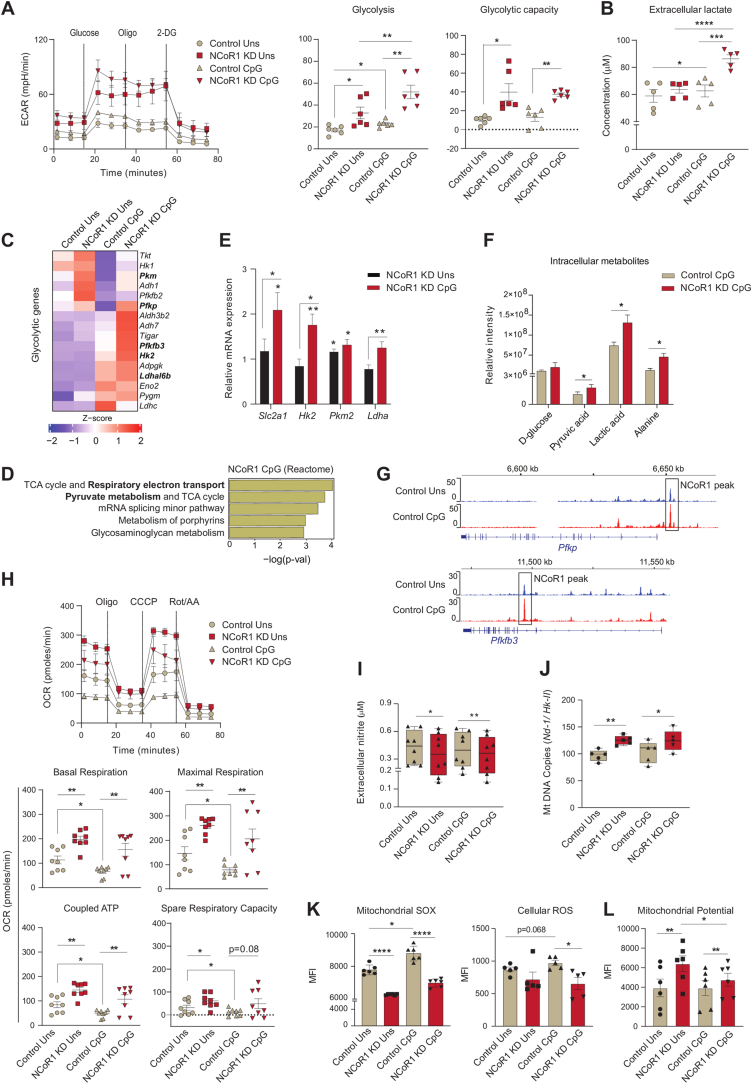
Fig. 1.
NCoR1 depleted tolerogenic cDC1 DCs depicted enhanced glycolysis and OXPHOS as compared to control cells
A. Representative line graph and scatter plots showing the glyco-stress parameters (glycolysis and glycolytic capacity) in unstimulated and 6h CpG activated control and NCoR1 depleted tolerogenic DCs, as measured using Seahorse extracellular flux analyzer. (n = 6)
B. Scatter plot depicting the extracellular lactate accumulation in the culture supernatants of unstimulated and 6h CpG activated control and NCoR1 depleted tolerogenic DCs at a dilution of 1:10. (n = 5)
C. Heatmap showing the differentially expressed genes (DEGs) enriched for the glycolytic pathway. RNAseq of unstimulated and 6h CpG activated NCoR1 KD as compared to control DCs were used to determine the DEGs. (n = 2)
D. Bar graphs depicting the top enriched metabolic pathways from the DEGs in NCoR1 KD CpG activated tolerogenic DCs as compared to controls, using Reactome Pathway Database. (n = 2)
E. RT-qPCR analysis showing the relative transcript expression of glut transporter Slc2a1, and glycolytic genes Hk2, Pkm2 and Ldha in unstimulated and 6h CpG activated NCoR1 KD tolerogenic DCs. (n = 5–7)
F. Bar graphs demostrating the relative intensities of intracellular metabolites of NCoR1 KD CpG activated DCs as compared to controls as quantified using GC-MS. (n = 3)
G. IGV snapshots showing the ChIPseq binding of NCoR1 at the Pfkp and Pfkfb3 gene loci in unstimulated and 6h CpG stimulated control DCs.
H. Representative line graph and scatter plots showing the mito-stress parameters (basal respiration, maximal respiration, coupled ATP and spare respiratory capacity) in unstimulated and 6h CpG activated control and NCoR1 depleted tolerogenic DCs as measured using an extracellular flux analyzer. (n = 8)
I. Box and whisker plots showing the min-max values for extracellular nitrite in the culture supernatant collected from unstimulated and 6h CpG activated control and NCoR1 depleted tolerogenic DCs. (n = 8)
J. Box and whisker plots depicting the min-max values for mitochondrial DNA copies (Nd-1/Hk-II) quantified by RT-qPCR in unstimulated and 6h CpG activated control and NCoR1 KD DCs. (n = 5)
K. Bar graphs with dots depicting the MFI levels of mitochondrial SOX, (n = 6) and cellular ROS, (n = 5) in unstimulated and 6h CpG stimulated control and NCoR1 KD DCs as quantified from flow cytometry.
L. Bar graphs with dots showing the MFI levels of TMRM measured for mitochondrial potential estimation in unstimulated and 6h CpG activated control and NCoR1 KD DCs. (n = 6)
*p ≤ 0.05, **p ≤ 0.01, ***p ≤0.001 and ****p ≤0.0001. p-value has been calculated using two tailed paired student’s t-test. Data shown in figure is combined from 3 independent experiments [A] and from 4 independent experiments [H]. Error bars represent SEM, except I and J, horizontal lines depicting mean.