FIGURE 6.
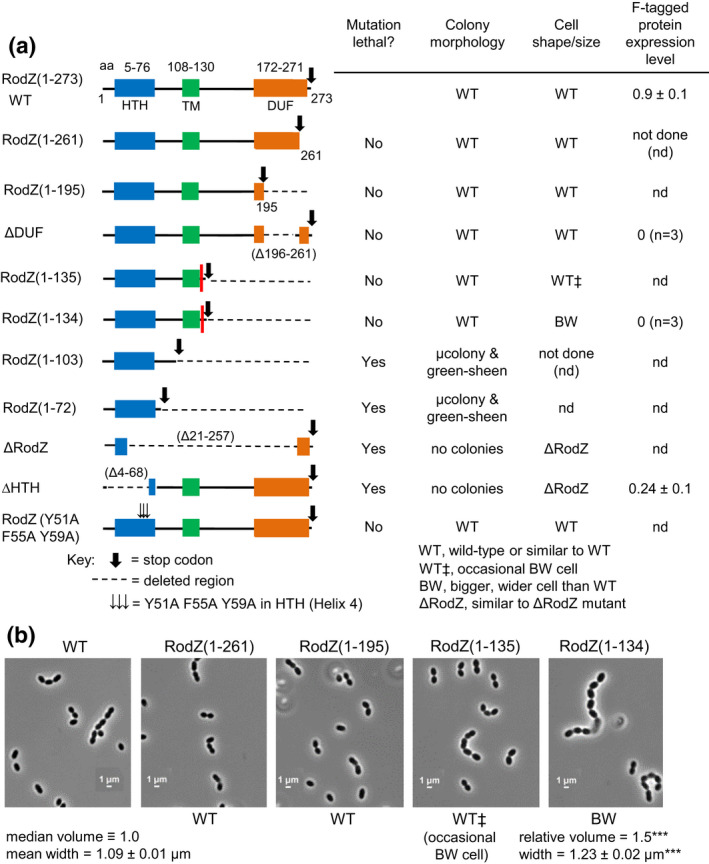
Amino acids 1–131 of RodZ are required for growth of Spn. (a) Amplicons harboring rodZ truncation or codon‐changing alleles were transformed into merodiploid strain IU12515 (ΔrodZ::Pc ‐[kan‐rpsL +] //PZn‐rodZ + ) to replace the Janus cassette (ΔrodZ::Pc ‐[kan‐rpsL +]) as described in experimental procedures and Table 2. Effects of RodZ truncations were determined by transformation assays on TSAII‐blood agar plates with or lacking Zn inducer (0.4 mM ZnCl2 + 0.04 mM MnSO4). Colony numbers, sizes, and morphologies were evaluated compared with rodZ + transformants after 20–24 h incubation at 37°C (see legend to Table 1 for experimental details). “μcolonies” (micro colonies) were barely visible by eye, but observed using a low power microscope. “Green‐sheen” refers to a shiny green pattern observed on top of the blood agar that may be due to partial hemolysis. Similar results were obtained in two independent transformation experiments (Table 2). The red bar between N131 and Y132 in the RodZ(1–135) and RodZ(1–134) entries marks the first TA site with a TnMariner insertion recovered by Tn‐seq of the WT strain (see Figure 2). Cell shapes and sizes were determined for WT and merodiploid mutants depleted for RodZ (see Figures 3 and S7; panel (b), below; Figures S8 and S9). Relative amounts of corresponding truncated RodZ proteins fused to a C‐terminus FLAG tag were determined by quantitative western blotting probed with anti‐FLAG antibody as described in Experimental procedures. Proteins samples were obtained from strains IU14594 (rodZ‐F at native chromosomal locus), IU13457 (rodZ‐F//PZn‐rodZ +), IU13655 (rodZ(ΔDUF)‐F//PZn‐rodZ +), IU13660 ((rodZ(1–134)‐F//PZn‐rodZ +), and IU13705 (rodZ(ΔHTH)‐F//PZn‐rodZ +) (see Table S1). Strains were grown in BHI broth +Zn inducer overnight, followed by growth for 4 h in BHI media lacking or containing Zn inducer as described in Figure 3. Values in the last column are amounts of truncated F‐tagged RodZ variants grown −Zn relative to the amount of RodZ‐F in IU14594. Although IU13655 (rodZ(ΔDUF)‐F//PZn‐rodZ +) and IU13660 ((rodZ(1–134)‐F//PZn‐rodZ +) were viable −Zn inducer, RodZ(ΔDUF)‐F and RodZ(1–134)‐F proteins were not detected in samples grown ± Zn, consistent with cleavage of the FLAG tag off the truncated RodZ variants lacking the C‐terminal DUF domain. (b) Representative micrographs of IU1824 (WT parent), and rodZ truncation mutants IU12794 (rodZ[1–261]//PZn‐rodZ +), IU12797 (rodZ[1–195]//PZn‐rodZ +), IU12799 (rodZ(1–135)//PZn‐rodZ +), and IU12803 (rodZ(1–134)//PZn‐rodZ +), which grow in the absence of Zn inducer. Cells were imaged during exponential growth at an OD620 ≈ 0.1–0.15 after ≈2.5–3.0 h of growth. Representative growth curves of truncated RodZ variants are shown in Figure S9d. Shapes and sizes were categorized as described for panel (a), above. Only the RodZ(M1‐Q134) mutant showed significant changes in relative median cell volume and average width (± SEM) compared with WT (n = 50 cells for each strain). ***, p < 0.001 by the non‐parametric, one‐way ANOVA Kruskal–Wallis test in GraphPad Prism. RodZ(M1‐T135) mutant cells resembled WT, except for an occasional bigger, wider cell.
