Abstract
Introduction
Analysis of measurable residual disease (MRD) is increasingly being implemented in the clinical care of children and adults with acute myeloid leukaemia (AML). However, MRD methodologies differ and discordances in results lead to difficulties in interpretation and clinical decision‐making. The aim of this study was to compare results from reverse transcription quantitative polymerase chain reaction (RT‐qPCR) and multiparameter flow cytometry (MFC) in childhood AML and describe the kinetics of residual leukaemic burden during induction treatment.
Methods
In 15 children who were treated in the NOPHO‐AML 2004 trial and had fusion transcripts quantified by RT‐qPCR, we compared MFC with RT‐qPCR for analysis of MRD during (day 15) and after induction therapy. Eight children had RUNX1::RUNX1T1, one CBFB::MYH11 and six KMT2A::MLLT3.
Results
When ≥0.1% was used as cut‐off for positivity, 10 of 22 samples were discordant. The majority (9/10) were MRD positive with RT‐qPCR but MRD negative with MFC, and several such cases showed the presence of mature myeloid cells. Fusion transcript expression was verified in mature cells as well as in CD34 expressing cells sorted from diagnostic samples.
Conclusions
Measurement with RT‐qPCR suggests slower response kinetics than indicated from MFC, presumably due to the presence of mature cells expressing fusion transcript. The prognostic impact of early measurements with RT‐qPCR remains to be determined.
Keywords: acute myeloid leukaemia, fusion transcript, measurable residual disease, multiparameter flow cytometry, RT‐qPCR
1. INTRODUCTION
Based on the results from several retrospective studies, analysis of measurable residual disease (MRD) is increasingly used for treatment allocation in patients with acute myeloid leukaemia (AML). However, MRD methodologies differ, and their results are not always concordant, resulting in difficulties in interpretation and clinical decision‐making. In children with de novo AML, MRD measured with multiparameter flow cytometry (MFC) after induction therapy is one of the strongest prognostic factors for both relapse and overall survival. 1 However, a substantial proportion of patients with undetectable MRD by MFC after induction relapses, as evidenced by a rate of almost 40% in MFC‐MRD negative patients in NOPHO‐AML 2004. Thus, MFC‐MRD cannot reliably identify low‐risk cases and there is a need for more sensitive measures of MRD, presumably using molecular methods. 1
Forty percent of children with AML have genetic aberrations in the leukaemic cells resulting in a quantifiable fusion transcript. 2 , 3 There are three major subgroups: the two core binding factor (CBF) leukaemias t(8;21)(q22;q22); RUNX1::RUNX1T1 and inv(16)(p13q22)/t(16;16)(p13;q22);CBFB::MYH11, and rearrangements of 11q23;KMT2Ar. 4 Reverse transcription followed by quantitative polymerase chain reaction (RT‐qPCR) gives a measurement of expression of leukaemia‐specific mRNA and enables a more sensitive analysis than MFC. In general, a sensitivity of at least 0.01% of the diagnostic fusion transcript level is obtained. 5 In adults with CBF‐AML, RT‐qPCR has a high prognostic value in identifying patients with a low relapse risk already after one induction course. 6 In childhood AML, only a few studies have evaluated the early treatment response with RT‐qPCR. The European Against Cancer program has defined appropriate reference genes and assays for RT‐qPCR, which has provided increased possibilities for standardization. 7 , 8 Using such assays, we have previously shown very high concordance between results with MFC and RT‐qPCR during induction therapy of ALL with t(12;21)(p13;q22); ETV6::RUNX1. 9 We have also shown the usefulness of RT‐qPCR for early detection of relapse in children with AML. 10
The aim of this study was to compare results from RT‐qPCR and MFC in childhood AML and describe the kinetics of residual leukaemic burden during induction treatment. We retrospectively analysed MFC and RT‐qPCR MRD results obtained at day 15 after start of the first induction course and before the first consolidation course in 15 children with de novo AML with RUNX1::RUNX1T1, CBFB::MYH11 or KMT2A::MLLT3 treated according to the NOPHO‐AML 2004 study protocol.
2. MATERIALS AND METHODS
2.1. Patients and samples
Fifteen patients with de novo AML with quantifiable fusion transcript and treated according to the NOPHO‐AML 2004 study protocol 11 were included: nine from Sweden, five from Denmark and one from Iceland. Patients were included when at least two of the following had been performed at diagnosis and day 15 after start of the first induction course and/or after the second induction course immediately before start of the first consolidation course (before consolidation, BC): MFC on bone marrow, RT‐qPCR of RUNX1::RUNX1T1/CBFB::MYH11/KMT2A::MLLT3 on blood and/or bone marrow. Patients and/or guardians consented to the study, which was performed according to the Declaration of Helsinki. The study was approved by the National Ethics Committees.
2.2. Quantitative polymerase chain reaction
RT‐qPCR was performed at National Reference Laboratories, Department of Clinical Chemistry, Sahlgrenska University Hospital for Swedish samples and at Haemodiagnostic Laboratory, Department of Haematology, Aarhus University Hospital for samples from Denmark and Iceland. RNA isolation and RT‐qPCR analyses for RUNX1::RUNX1T1, CBFB::MYH11, and KMT2A::MLLT3 were performed as previously described 10 with the Swedish reference laboratory labelled lab no V and the Danish reference laboratory lab no I, in large according to the recommendations from the EAC program. 7 , 8 For quantification of MRD, the Swedish reference laboratory used GUSB as reference gene and ratios of copy numbers were normalized to the diagnostic level and the Danish reference laboratory used the comparative CT (ΔΔCq) method with B2M and ABL1 as reference genes. 12 The cut‐off level for fusion transcript MRD positivity was set to ≥0.1% of diagnostic level.
2.3. Multiparameter flow cytometry
Multiparameter flow cytometry files from day 15 after the first induction course and/or before the first consolidation course were available from 13 patients. The MFC analyses were performed in two of the Nordic laboratories in Sweden (Gothenburg) and Denmark (Copenhagen), both members of the Nordic Flow Cytometry Group. The analyses were performed on FacsCalibur or FacsCanto instruments using the FacsDiva software (Becton Dickenson, Erembodegem, Belgium). The 4–8 antibody combinations used were at the discretion of each laboratory, but contained CD2, CD5, CD7, CD4, CD19, CD11b, CD11c, CD13, CD14, CD15, CD16, CD33, CD34, CD36, CD38, CD41, CD42b, CD56, CD61, CD64, CD65, CD99, CD117, CD123, CD133, CD135 and HLA‐DR. Data files were reviewed using the leukaemia associated immunophenotype (LAIP) approach as previously described. 1 The MFC‐MRD level was expressed as percentage of leukaemic cells of all viable cells. A minimum of 100 clustered leukaemic events was required for diagnosis of residual disease. The cut‐off level for MFC‐MRD positivity was set to ≥0.1% leukaemic cells. Analysis of 100 000 viable cells were required to score a sample as MFC‐MRD negative. For day 15 samples, MFC data files were also assessed for other cell types, as possible based on antibody combinations used.
2.4. Cell sorting
For sorting of subpopulations with fluorescence‐activated cell sorting, viably frozen bone marrow cells from diagnostic samples were used. Cells were thawed, washed and stained with PerCP‐Cy5.5‐conjugated anti‐CD34 antibody, Horizon V450‐conjugated anti‐HLA‐DR, Horizon V500‐conjugated anti‐CD45 (all Becton Dickinson, San Jose, CA), and PE‐Cy7‐conjugated anti‐CD117 (Beckman Coulter, Brea, CA). Sorting of the following cell populations was performed on an FACSAria (Becton Dickinson): CD34+CD117+ cells, CD34‐CD117+ cells, granulocytes (mainly neutrophils) based on CD45+, high SSC, CD34‐CD117‐, lymphocytes based on CD45++, low SSC, CD34‐CD117‐, and when applicable mast cells based on CD34‐, CD117++. Sorted cells were subjected to RT‐qPCR as described above, with determination of ratios of copy numbers and ABL1 as reference gene.
2.5. Statistical analyses
IBM SPSS Statistics for MAC, Version 21 (IBM Corp., Armonk, NY) was used for statistical analyses. Concordance of MRD results was assessed with Cohen's Kappa (K) test and correlations with Spearman's rank correlation test. All tests were two‐sided, and P < 0.05 was considered significant.
3. RESULTS
3.1. Patient characteristics
The study included 15 children (8 females and 7 males, median age 6 years (range: 1–16), diagnosed 2004–2011 with de novo AML with a quantifiable fusion transcript (8 with RUNX1::RUNX1T1, one with CBFB::MYH11 and 6 with KMT2A::MLLT3). At diagnosis, all patients had white blood cell counts less than 100 × 109/L (median 22.5 x109/L, range 4.7–98.2 × 109/L), two had CNS involvement and two had extramedullary disease. All patients achieved complete remission. Nine of the patients (five with RUNX1::RUNX1T1 and four with KMT2A::MLLT3) experienced relapse (median time from diagnosis 12 months, range: 8–16 months). Of the patients with relapse, six died. The median follow‐up time for the patients that remained alive was 76 months (range: 38–91 months).
3.2. Kinetics of fusion transcripts during induction
In order to assess the reduction of fusion transcript levels during treatment, RT‐qPCR was performed shortly after the first cycle, that is, at day 15, and at the last bone marrow examination before start of consolidation (BC). The kinetics of reduction of fusion transcripts assessed with RT‐qPCR are shown for individual patients in Figure 1 and for groups as log reductions in Figure 2A. At day 15, 11/12 analysed patients had quantifiable fusion transcripts in bone marrow and one patient had detectable but not quantifiable fusion transcripts. In blood, 8/10 patients had quantifiable levels and two patients had detectable but not quantifiable levels. At the time point BC, 8/15 patients had quantifiable levels in bone marrow, five patients had detectable but not quantifiable levels, and the remaining two patients had no detectable fusion transcripts. In blood, 6/13 patients had quantifiable levels, four patients had detectable but not quantifiable, and three patients had no detectable fusion transcript. When we compared fusion transcript levels in bone marrow and blood at both these time points, there was a significant correlation (r s = 0.93; P < 0.001, n = 22). When dichotomizing results into MRD positive and MRD negative based on the cut‐off 0.1%, there was a total 91% (20/22 samples) agreement between bone marrow and blood (K‐value 0.82; P < 0.001). The two discordant results were from BC and displayed positive RT‐qPCR in bone marrow but not in blood. In summary, bone marrow and blood showed the similar results.
FIGURE 1.
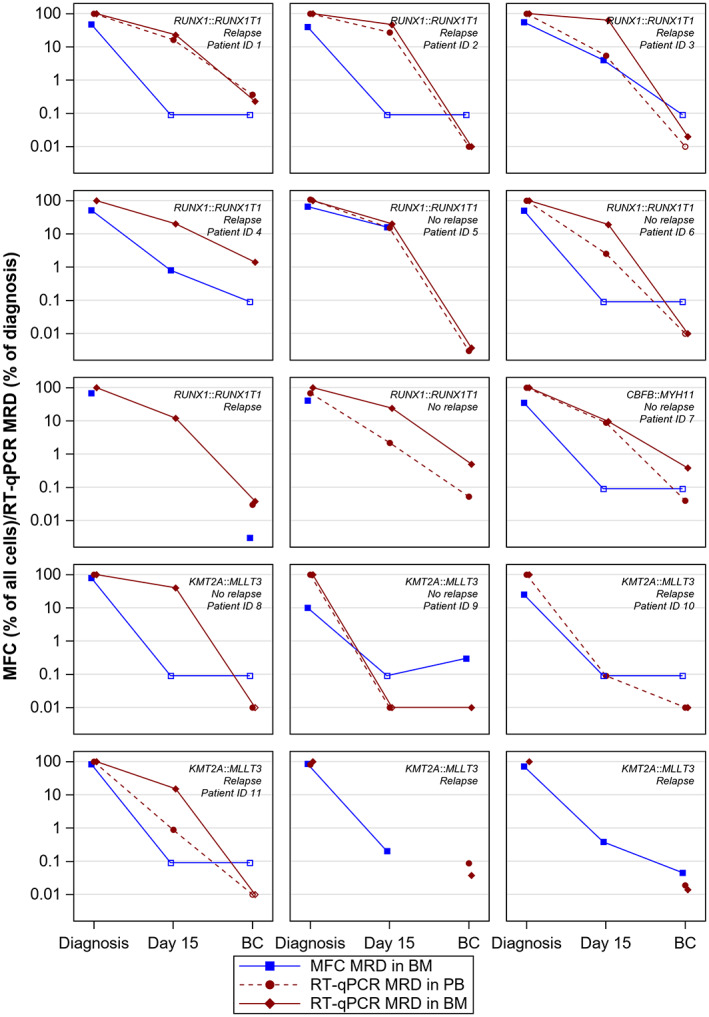
The kinetics of MRD during treatment in 15 children with AML. Fusion transcripts were assessed with RT‐qPCR in bone marrow and peripheral blood and MRD analysis with MFC was performed in bone marrow. Measurements were performed at diagnosis, day 15 in treatment and before start of consolidation (BC). Genetic aberration (RUNX1::RUNX1T1/CBFB::MYH11/KMT2A::MLLT3), relapse status, and ID (only applicable to patients depicted in Figure 2B) are stated. Filled symbols represent detectable levels of MRD and unfilled undetectable levels (depicted at level of detection for MFC and level of quantification for RT‐qPCR).
FIGURE 2.
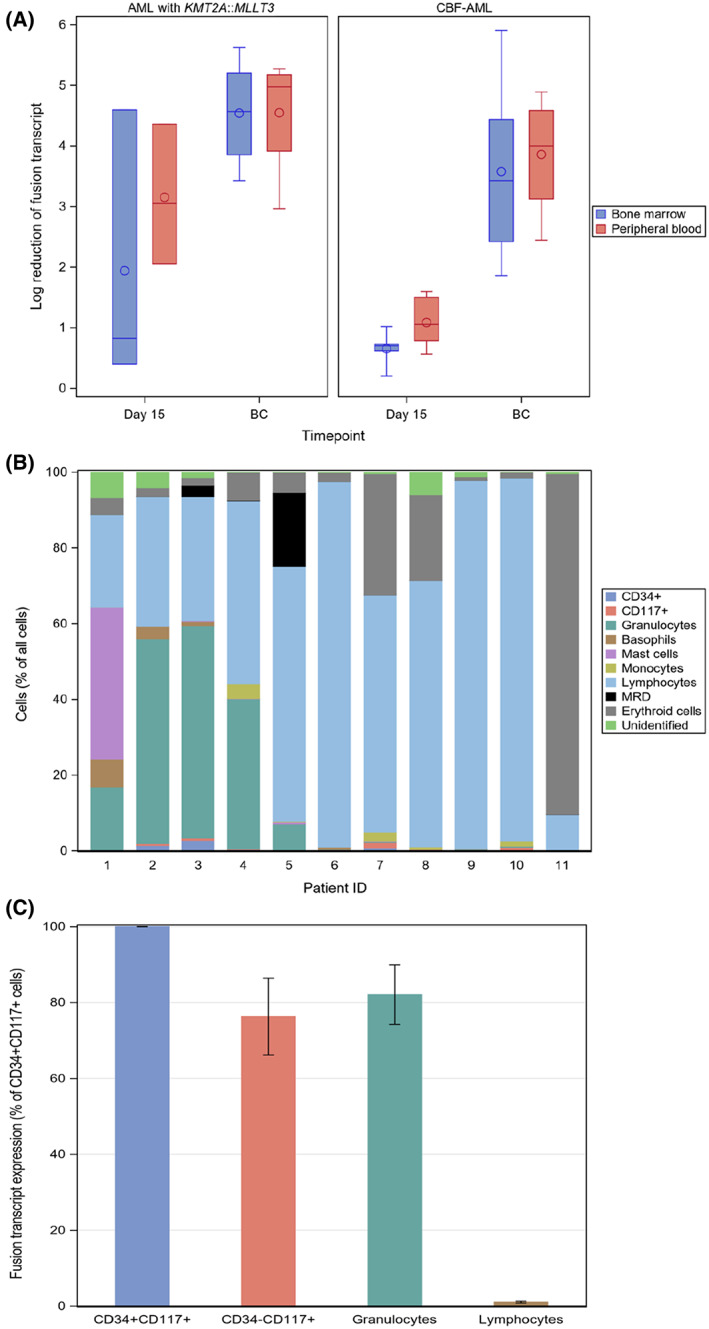
A. Box plots showing fusion transcript log reductions at day 15 and before start of consolidation (BC) in bone marrow and peripheral blood. Cases with RUNX1::RUNX1T1 or CBFB::MYH11, combined as core binding factor AML (CBF‐AML), and cases with AML with KMT2A::MLLT3 are shown. In the boxes, circles are mean values and horizontal lines are medians. B. Bone marrow cell content assessed with MFC day 15 in 11 children with AML. ID:s are the same as in Figure 2 with ID 1–6 being AML with RUNX1::RUNX1T1, ID 7 AML with CBFB::MYH11, and ID 8–11 AML with KMT2A::MLLT3; ID 1–4 and 10–11 relapsed, ID 5–9 did not experience relapse. Cells were identified based on the antibody combinations used for each case in the MRD analysis. In the graph, ‘CD34+’ cells included CD117 and CD34 double positive cells, ‘CD117+’ cells included only CD34‐cells, and ‘unidentified’ cells could not be reliable identified based on the antibody combinations used. C. The mRNA expression of fusion transcript RUNX1::RUNX1T1 (n = 5) or CBFB::MYH11 (n = 2) in CD34+CD117+ cells, CD34‐CD117+ cells, granulocytes and lymphocytes. Cells were sorted from viably frozen diagnostic bone marrow samples from five children and two adults with CBF AML.
3.3. Discordance between bone marrow MRD levels with MFC and RT‐qPCR
To assess the concordance between RT‐qPCR and MFC for MRD detection, we performed correlation and agreement analyses on results from bone marrow samples. When MFC and RT‐qPCR results from day 15 and BC were analysed, there was no correlation between MFC and RT‐qPCR (r s = 0.32: P = 0.15, and n = 22). We also performed agreement analyses between RT‐qPCR MRD and MFC MRD using the cut‐off level 0.1% for both methods. Only 12 of 22 samples were concordant (4/10 at day 15, 8/12 BC, Table 1). Most discordant results (9/10) showed positivity (≥0.1%) with RT‐qPCR but negativity (<0.1%) with MFC. There was the similar level of discordance (11/22) also when it was defined as >1 log10 difference between levels obtained with RT‐qPCR an MFC. The similar trend was seen when comparing MFC with RT‐qPCR in blood (Table 1). This pattern of slower response kinetics with higher MRD results obtained with RT‐qPCR was seen in all subtypes and most evidently in cases with RUNX1::RUNX1T1 (Figure 1). At the time point BC, there was a 67% concordance between MFC and RT‐qPCR in bone marrow (Table 1). Among the four samples with discordant result, RT‐qPCR MRD was ≥0.1% in three cases where flow cytometry MRD was <0.1%. Of these, two, both with RUNX1::RUNX1T1, experienced relapse and died. The third had AML with CBFB::MYH11 and did not relapse. The fourth discordant case had AML with KMT2A::MLLT3 where the RT‐qPCR showed detectable but not quantifiable fusion transcript and MFC MRD was positive, this patient did not relapse. In conclusion, RT‐qPCR often showed more residual disease than MFC, particularly in CBF‐AML.
TABLE 1.
Concordance between MRD results by RT‐qPCR and MFC
| MFC MRD | RT‐qPCR MRD in bone marrow | RT‐qPCR MRD in peripheral blood | ||
| Positive (≥0.1%) | Negative (<0.1%) | Positive (≥0.1%) | Negative (<0.1%) | |
| All time points | ||||
| Positive (≥0.1%) | 3 | 1 | 2 | 0 |
| Negative (<0.1%) | 9 | 9 | 6 | 11 |
| Concordance | 54% | 68% | ||
| K‐value | 0.141 (p = 0.364) | 0.278 (p = 0.080) | ||
| No. samples | 22 | 19 | ||
| Day 15 | ||||
| Positive (≥0.1%) | 3 | 0 | 2 | 0 |
| Negative (<0.1%) | 6 | 1 | 5 | 2 |
| Concordance | 40% | 44% | ||
| K‐value | 0.091 (p = 0.490) | 0.151 (p = 0.391) | ||
| No. samples | 10 | 9 | ||
| BC | ||||
| Positive (≥0.1%) | 0 | 1 | 0 | 0 |
| Negative (<0.1%) | 3 | 8 | 1 | 9 |
| Concordance | 67% | 90% | ||
| K‐value | −0.143 (p = 0.546) | |||
| No. samples | 12 | 10 | ||
Abbreviations: MFC, multiparameter flow cytometry; MRD, measurable residual disease; RT‐qPCR, real‐time quantitative polymerase chain reaction.
3.4. Bone marrow cell populations at day 15
Because of the observed discordance in kinetics between MFC and RT‐qPCR, we reanalyzed MFC data files from day 15 with the purpose to describe the cell populations present in the samples (Figure 2B). In several cases, and most evident in AML with RUNX1::RUNX1T1, the bone marrow contained mature cell types such as granulocytes, mast cells, basophils, CD34‐CD117+ cells and/or monocytes. Among the three discordant RUNX1::RUNX1T1 cases (Figures 1 and 2B, one (ID 1) showed substantial amounts of mast cells, basophils and granulocytes, but no evidence of cells with leukaemia associated immunophenotype (LAIP). Another case (ID 2) showed the presence of basophils and granulocytes and a low frequency of CD34+CD117+ cells (1.4%) not expressing the LAIP, which in this case was CD56 positivity in a subpopulation of CD34+CD117+ cells. Both these cases relapsed. The discordant RUNX1::RUNX1T1 case (ID 6) who did not relapse contained almost exclusively lymphocytes at day 15. The only case with CBFB::MYH11 (ID 7, no relapse) showed mostly lymphocytes, but with rather high amounts of erythroid cells, monocytes and granulocytes, and the presence of CD34‐CD117+ cells without LAIP. In patients with KMT2A::MLLT3 (ID 8–11), day 15 bone marrow contained mostly lymphocytes. In one of these cases, ID 11 who eventually relapsed, there was a substantial amount of erythroid cells, which influenced the MRD level with MFC (0.07% of viable cells) and most probably explained the discordance with negative MFC and clearly positive RT‐qPCR. In conclusion, the most prominent finding was the presence of mature cell types in discordant cases of CBF‐AML.
3.5. Fusion transcripts are present in both immature and mature myeloid cells at diagnosis of CBF‐AML
Since several of the CBF‐AML cases with remaining fusion transcript expression showed the presence of mature cells at day 15, we hypothesised that such cells could express the fusion transcripts. We therefore determined the expression of fusion transcripts in mature cell populations in bone marrow samples obtained at diagnosis from five of the children included in this study and from two adult patients (Figure 2C). The following cell populations were sorted and analysed for transcript levels of RUNX1::RUNX1T1 (n = 5) or CBFB::MYH11 (n = 2): CD34+CD117+ cells, CD34‐CD117+ cells, granulocytes, mast cells (when present), and lymphocytes as negative control. Leukaemic transcripts were clearly present in CD34+CD117+ cells, CD34‐CD117+ cells and granulocytes, but not in lymphocytes above what was considered as contamination from sorting. In one case with RUNX1::RUNX1T1 and one with CBFB::MYH11, mast cells showing the expression of fusion transcript (at levels of 0.2 and 2 ‐fold relative to the expression in CD34+CD117+ cells, respectively) were present in the diagnostic samples. Thus, fusion transcripts were present in both immature and mature myeloid cells at diagnosis of CBF‐AML.
4. DISCUSSION
In this study, we compared MFC with the expectedly more sensitive method RT‐qPCR for quantification of MRD during and after induction therapy. We found a low concordance between RT‐qPCR and MFC in bone marrow during and after induction treatment, with a pattern of slower response kinetics for fusion transcripts than leukaemic cells as determined by MFC, especially in cases with CBF‐AML. This might be due to the presence of fusion transcripts in cell types not defined as MRD by MFC due to their mature antigen expression.
The prognostic impact of the early treatment response measured with MFC in children with de novo AML is well established and may be applicable for the majority of children. 1 , 13 , 14 Nevertheless, MFC fails to identify a substantial proportion of patients that relapse. When we compared RT‐qPCR with MFC in a subset of patients included in the NOPHO‐AML 2004 trial in which MFC‐MRD has shown independent prognostic impact at day 15 and before consolidation, we found a low concordance between the methods. RT‐qPCR revealed a pattern of slower treatment kinetics than MFC during induction therapy, especially in cases with CBF‐AML. In most of the discordant samples in our study, MRD could only be detected by RT‐qPCR and not with MFC. This was not explained by different materials used for the assays, since all MFC analyses and the majority of RT‐qPCR analyses were performed on unfractionated bone marrow. Similar results have been reported in both children 15 and young adults 16 but with contradictory conclusions regarding the value of RT‐qPCR in CBF‐AML. In 65 young adults with CBF‐AML and 2‐year leukaemia‐free survival of 73% and 50% for RUNX1::RUNX1T1 and CBFB::MYH11 respectively, Perea et al noted an equally strong prognostic value for MFC‐MRD and RT‐qPCR with a superior prognosis for patients with undetectable MRD with both methods. 16 Inaba et al, on the other hand, found a prognostic value for MFC‐MRD but not of RT‐qPCR MRD levels in a paediatric cohort of CBF‐AML. 15 Of note, this cohort had an excellent relapse‐free survival (only 3 out of 55 patients relapsed), and qPCR analysis was performed using GAPDH as reference gene, i.e. not according to recommendations from the Europe Against Cancer program, as used in our study and by Perea et al. 16 Zhang et al. reported the prognostic value of RT‐qPCR (with ABL1 as reference gene) after induction as well as during consolidation in a cohort of children with AML with t(8;21)(q22;22) in which 14 of 62 experienced relapsed. 17 In the trial in which our study cohort was treated, NOPHO‐AML 2004, the results for AML with RUNX1::RUNX1T1 were poorer than expected with an EFS of only 35%. 18 The patients studied herein were thus representative of this particular trial. This high frequency of relapses implies a poor efficacy of induction therapy, which might be one reason for high fusion transcript levels. In summary, both technical issues and treatment efficacy may affect the results of studies comparing MRD methods. To establish the clinical value of MRD methods, the methodologies must be carefully evaluated.
In order to find the reason for the remaining high levels of fusion transcripts day 15, we reanalyzed flow cytometry files aiming at identifying all cell types present. These analyses confirmed the absence of immature cells with LAIP. In principle, we observed two different patterns: (1) the presence of mature myeloid cells and a relatively good cellularity, or (2) high proportion of lymphocytes and presence of immature cells without LAIP in hypoplastic bone marrow. Based on the first pattern we hypothesised that fusion transcripts are present in mature cells not defined as MRD by MFC. Such cells lack immature markers, and presumably leukemogenic potential, and are therefore usually not scored as MRD. We confirmed expression of fusion transcripts in mature cells using sorted immature and mature cells from diagnostic bone marrow samples from patients with CBF‐AML. We detected high levels of fusion transcript in CD34‐CD117+ progenitors, mature granulocytes (CD34‐CD117‐) and mast cells, as well as in the most immature CD34+CD117+ progenitors. The patient displaying high amounts of mast cells day 15 in our study had RUNX1::RUNX1T1 expressing mast cells at diagnosis. Mast cells have previously been reported to have the potential to harbour leukaemic fusions. 19 The expression of fusion transcripts in mature myeloid cells with unknown leukemogenic potential is a common finding in APL, and probably the reason for the lack of prognostic value of fusion transcripts early in the treatment of APL. 20 The similar finding, albeit the presence of mutations, not transcripts, has been observed during treatment of AML with IDH inhibitors. 21 , 22 Therefore, the interpretation of results from RT‐qPCR and other molecular analyses in the evaluation of an early treatment response is not obvious. In our study, this is illustrated by three relapses among the six patients that at day 15 had positive MRD by RT‐qPCR but negative by MFC. Several of the cases with high levels of RUNX1::RUNX1T1 eventually relapsed, a pattern not obvious in the fewer cases with KMT2A::MLLT3. However, it must be emphasized that this study is too small to draw clinical conclusions. Thus, further studies are needed to determine the prognostic impact of early measurements with RT‐qPCR and its head‐to‐head comparison to MFC‐MRD, as is now ongoing in the paediatric trial NOPHO‐DBH AML 2012 (EudraCT 2012‐002934‐35). In parallel, RT‐qPCR has been included for risk stratification in at least one childhood AML protocol, MyeChild 01 (EudraCT 2014–005066‐30).
Discordance between RT‐qPCR and MFC was in our study observed both day 15 and BC. We, and others, have demonstrated that fusion transcripts can be detected by RT‐qPCR in BM also late after clinical remission is achieved. 3 , 10 , 23 , 24 , 25 , 26 This has limited the use of RT‐qPCR and may have several explanations in addition to our hypothesis regarding fusion transcript expression in mature cells. First, RT‐qPCR is a more sensitive method than MFC, at least for RUNX1::RUNX1T1 and CBFB::MYH11 and might therefore detect residual disease at a lower level. Second, there is no direct relationship between the number of fusion transcripts and the number of cells they originate from, that is, the same level of fusion transcript may arise from a few high‐expressing cells or from many low‐expressing cells. Whether there is a relationship between expression of fusion transcripts and leukaemia‐initiating capacity on the single‐cell level is not known. Third, longstanding fusion transcript positivity in BM may result from the presence of preleukemic cells containing the fusion gene but not secondary mutations. 27 Finally, a fourth possibility is that leukaemic cells are in fact present in the bone marrow but cannot be distinguished from normal myoblasts based on the available MFC MRD technique. 26 , 27 In our series, there was a very strong concordance between levels of fusion transcripts in blood and bone marrow, probably since all samples were from early time points in treatment. When measured during consolidation or after end of treatment, transcripts can be detected in bone marrow without clinical implication, while the presence of fusion transcripts in blood is a very strong predictor of relapse. 10
In conclusion, during and after induction therapy of childhood AML, RT‐qPCR has a higher propensity to detect remaining leukaemic cells than MFC. Whether this is clinically relevant remains to be shown and is now being investigated in the NOPHO‐DBH AML 2012 trial.
AUTHOR CONTRIBUTIONS
Lene Karlsson, Linda Fogelstrand and Jonas Abrahamsson designed the study. Lene Karlsson, Charlotte Guldborg Nyvold, Anastasia Soboli, Pegah Johansson, Lars Palmqvist, Anne Tierens, Henrik Hasle, Birgitte Lausen, Ólafur Gisli Jónsson, Gitte Wulff Jürgensen, Lene Hyldahl Ebbesen, and Jonas Abrahamsson contributed data. Lene Karlsson, Anastasia Soboli, Linda Fogelstrand and Jonas Abrahamsson analysed data and wrote the manuscript. All authors reviewed the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGEMENTS
We thank the employees at the Department of Clinical Chemistry at Sahlgrenska University Hospital, Haemodiagnostic Laboratory at the Aarhus University Hospital, and Department of Clinical Immunology, Copenhagen University Hospital Rigshospitalet for sample collection, processing and analyses.
Karlsson L, Nyvold CG, Soboli A, et al. Fusion transcript analysis reveals slower response kinetics than multiparameter flow cytometry in childhood acute myeloid leukaemia. Int J Lab Hematol. 2022;44(6):1094‐1101. doi: 10.1111/ijlh.13935
Jonas Abrahamsson and Linda Fogelstrand contributed equally to the study.
Funding information The Swedish Childhood Cancer fund; The Swedish state under the agreement between the Swedish government and the county councils, the ALF‐agreement, Grant/Award Numbers: ALFGBG‐720681, ALFGBG‐70550
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Tierens A, Bjorklund E, Siitonen S, et al. Residual disease detected by flow cytometry is an independent predictor of survival in childhood acute myeloid leukaemia; results of the NOPHO‐AML 2004 study. Br J Haematol. 2016;174(4):600‐609. [DOI] [PubMed] [Google Scholar]
- 2. Viehmann S, Teigler‐Schlegel A, Bruch J, Langebrake C, Reinhardt D, Harbott J. Monitoring of minimal residual disease (MRD) by real‐time quantitative reverse transcription PCR (RQ‐RT‐PCR) in childhood acute myeloid leukemia with AML1/ETO rearrangement. Leukemia. 2003;17(6):1130‐1136. [DOI] [PubMed] [Google Scholar]
- 3. Matsuo H, Iijima‐Yamashita Y, Yamada M, et al. Monitoring of fusion gene transcripts to predict relapse in pediatric acute myeloid leukemia. Pediatr Int: Off J Japan Pediatr Soc. 2018;60(1):41‐46. [DOI] [PubMed] [Google Scholar]
- 4. Creutzig U, Zimmermann M, Reinhardt D, et al. Changes in cytogenetics and molecular genetics in acute myeloid leukemia from childhood to adult age groups. Cancer. 2016;122(24):3821‐3830. [DOI] [PubMed] [Google Scholar]
- 5. Ommen HB. Monitoring minimal residual disease in acute myeloid leukaemia: a review of the current evolving strategies. Ther Adv Hematol. 2016;7(1):3‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yin JA, O'Brien MA, Hills RK, et al. Minimal residual disease monitoring by quantitative RT‐PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML‐15 trial. Blood. 2012;120(14):2826‐2835. [DOI] [PubMed] [Google Scholar]
- 7. Beillard E, Pallisgaard N, van der Velden VH, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using 'real‐time' quantitative reverse‐transcriptase polymerase chain reaction (RQ‐PCR) ‐ a Europe against cancer program. Leukemia. 2003;17(12):2474‐2486. [DOI] [PubMed] [Google Scholar]
- 8. Gabert J, Beillard E, van der Velden VH, et al. Standardization and quality control studies of 'real‐time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia – a Europe against cancer program. Leukemia. 2003;17(12):2318‐2357. [DOI] [PubMed] [Google Scholar]
- 9. Alm SJ, Engvall C, Asp J, Palmqvist L, Abrahamsson J, Fogelstrand L. Minimal residual disease monitoring in childhood B lymphoblastic leukemia with t(12;21)(p13;q22); ETV6‐RUNX1: concordant results using quantitation of fusion transcript and flow cytometry. Int J Lab Hematol. 2017;39(2):121‐128. [DOI] [PubMed] [Google Scholar]
- 10. Juul‐Dam KL, Ommen HB, Nyvold CG, et al. Measurable residual disease assessment by qPCR in peripheral blood is an informative tool for disease surveillance in childhood acute myeloid leukaemia. Br J Haematol. 2020;190:198‐208. [DOI] [PubMed] [Google Scholar]
- 11. Abrahamsson J, Forestier E, Heldrup J, et al. Response‐guided induction therapy in pediatric acute myeloid leukemia with excellent remission rate. J Clin Oncol. 2011;29(3):310‐315. [DOI] [PubMed] [Google Scholar]
- 12. van der Velden VH, Hochhaus A, Cazzaniga G, et al. Detection of minimal residual disease in hematologic malignancies by real‐time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia. 2003;17(6):1013‐1034. [DOI] [PubMed] [Google Scholar]
- 13. Buldini B, Rizzati F, Masetti R, et al. Prognostic significance of flow‐cytometry evaluation of minimal residual disease in children with acute myeloid leukaemia treated according to the AIEOP‐AML 2002/01 study protocol. Br J Haematol. 2017;177(1):116‐126. [DOI] [PubMed] [Google Scholar]
- 14. Rubnitz JE, Inaba H, Dahl G, et al. Minimal residual disease‐directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6):543‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inaba H, Coustan‐Smith E, Cao X, et al. Comparative analysis of different approaches to measure treatment response in acute myeloid leukemia. J Clin Oncol. 2012;30(29):3625‐3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perea G, Lasa A, Aventin A, et al. Prognostic value of minimal residual disease (MRD) in acute myeloid leukemia (AML) with favorable cytogenetics [t(8;21) and inv(16)]. Leukemia. 2006. Jan;20(1):87‐94. [DOI] [PubMed] [Google Scholar]
- 17. Zhang L, Cao Z, Ruan M, et al. Monitoring the AML1/ETO fusion transcript to predict outcome in childhood acute myeloid leukemia. Pediatr Blood Cancer. 2014;61(10):1761‐1766. [DOI] [PubMed] [Google Scholar]
- 18. Hasle H, Abrahamsson J, de Bont E, et al. Anthracycline type during induction associated with outcome in pediatric t(8;21) and Inv(16) AML [meeting abstract]. Blood. 2014;124(21):11.25121166 [Google Scholar]
- 19. Pullarkat V, Bedell V, Kim Y, et al. Neoplastic mast cells in systemic mastocytosis associated with t(8;21) acute myeloid leukemia are derived from the leukemic clone. Leuk Res. 2007;31(2):261‐265. [DOI] [PubMed] [Google Scholar]
- 20. Grimwade D, Jovanovic JV, Hills RK, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre‐emptive arsenic trioxide therapy. J Clin Oncol. 2009;27(22):3650‐3658. [DOI] [PubMed] [Google Scholar]
- 21. Ragon BK, DiNardo CD. Targeting IDH1 and IDH2 mutations in acute myeloid leukemia. Curr Hematol Malig Rep. 2017;512(6):537‐546. [DOI] [PubMed] [Google Scholar]
- 22. Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722‐731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buonamici S, Ottaviani E, Testoni N, et al. Real‐time quantitation of minimal residual disease in inv(16)‐positive acute myeloid leukemia may indicate risk for clinical relapse and may identify patients in a curable state. Blood. 2002;99(2):443‐449. [DOI] [PubMed] [Google Scholar]
- 24. Leroy H, de Botton S, Grardel‐Duflos N, et al. Prognostic value of real‐time quantitative PCR (RQ‐PCR) in AML with t(8;21). Leukemia. 2005;19(3):367‐372. [DOI] [PubMed] [Google Scholar]
- 25. Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long‐term remission. Blood. 1993;82(3):712‐715. [PubMed] [Google Scholar]
- 26. Zhou Y, Wood BL. Methods of detection of measurable residual disease in AML. Curr Hematol Malig Rep. 2017;2:557‐567. [DOI] [PubMed] [Google Scholar]
- 27. Christen F, Hoyer K, Yoshida K, et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients. Blood. 2019;133(10):1140‐1151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


