Abstract
Background
Radiomics has been considered an imaging marker for capturing quantitative image information (QII). The introduction of radiomics to image segmentation is desirable but challenging.
Purpose
This study aims to develop and validate a radiomics‐based framework for image segmentation (RFIS).
Methods
RFIS is designed using features extracted from volume (svfeatures) created by sliding window (swvolume). The 53 svfeatures are extracted from 11 phantom series. Outliers in the svfeature datasets are detected by isolation forest (iForest) and specified as the mean value. The percentage coefficient of variation (%COV) is calculated to evaluate the reproducibility of svfeatures. RFIS is constructed and applied to the gross target volume (GTV) segmentation from the peritumoral region (GTV with a 10 mm margin) to assess its feasibility. The 127 lung cancer images are enrolled. The test–retest method, correlation matrix, and Mann–Whitney U test (p < 0.05) are used to select non‐redundant svfeatures of statistical significance from the reproducible svfeatures. The synthetic minority over‐sampling technique is utilized to balance the minority group in the training sets. The support vector machine is employed for RFIS construction, which is tuned in the training set using 10‐fold stratified cross‐validation and then evaluated in the test sets. The swvolumes with the consistent classification results are grouped and merged. Mode filtering is performed to remove very small subvolumes and create relatively large regions of completely uniform character. In addition, RFIS performance is evaluated by the area under the receiver operating characteristic (ROC) curve (AUC), accuracy, sensitivity, specificity, and Dice similarity coefficient (DSC).
Results
30249 phantom and 145008 patient image swvolumes were analyzed. Forty‐nine (92.45% of 53) svfeatures represented excellent reproducibility(%COV<15). Forty‐five features (91.84% of 49) included five categories that passed test‐retest analysis. Thirteen svfeatures (28.89% of 45) svfeatures were selected for RFIS construction. RFIS showed an average (95% confidence interval) sensitivity of 0.848 (95% CI:0.844–0.883), a specificity of 0.821 (95% CI: 0.818–0.825), an accuracy of 83.48% (95% CI: 83.27%–83.70%), and an AUC of 0.906 (95% CI: 0.904–0.908) with cross‐validation. The sensitivity, specificity, accuracy, and AUC were equal to 0.762 (95% CI: 0.754–0.770), 0.840 (95% CI: 0.837–0.844), 82.29% (95% CI: 81.90%–82.60%), and 0.877 (95% CI: 0.873–0.881) in the test set, respectively. GTV was segmented by grouping and merging swvolume with identical classification results. The mean DSC after mode filtering was 0.707 ± 0.093 in the training sets and 0.688 ± 0.072 in the test sets.
Conclusion
Reproducible svfeatures can capture the differences in QII among swvolumes. RFIS can be applied to swvolume classification, which achieves image segmentation by grouping and merging the swvolume with similar QII.
Keywords: computed tomography, image segmentation, radiomics, tumor
1. INTRODUCTION
Radiomics provide quantitative information on medical imaging data by extracting and analyzing quantitative imaging features 1 and can hold predictive information to guide personalized radiotherapy (RT). 2 The application of radiomics to image segmentation is of great interest since it may contribute to designing more personalized RT plans, thereby reducing radiation toxicity.
Segmentation of computed tomography (CT) images is a critical step in RT plan design, 3 which is usually carried out manually by radiation oncologists following recommended guidelines. Considering the low soft tissue contrast, CT‐based image segmentation relying on visual assessment remains challenging for radiation oncologists. 4 , 5 . Deep‐learning (DL) technology has been applied to medical image processing, and Unet‐based models have achieved remarkable success in medical image segmentation tasks. 6 However, the great performance of end‐to‐end DL networks comes at the cost of high complexity and numerous parameters. We could not explain why certain classifications were made in image segmentation in DL networks. The final outputs of DL networks are accepted without justification, reducing physicians’ confidence in DL network applications.
In addition, the training time and limited dataset also pose challenges to the application of DL networks in image segmentation. Numerous parameters result in the time‐consuming training of DL‐based segmentation networks. Due to cost constraints and privacy protection, it is intractable to acquire mass medical images for the DL network. 7 Moreover, advanced models may underperform simple models if data samples change. 6 Simple and efficient unsupervised clustering algorithms have been used for image segmentation, such as the K‐means clustering algorithm. 8 , 9 However, identifying the biological and/or clinical significance of the volume segmented by unsupervised clustering algorithms is still troublesome. For this reason, it is under urgent demand to develop a simple and supervised medical image segmentation model with ideal generalization.
Radiomics, capable of capturing the quantitative information within medical images, have been applied in medical image analysis. 10 Radiomic features provide objective and quantitative methods for tumor phenotype assessment and enjoy widespread potential applications in oncology. 11 , 12 For example, radiomic features have shown promise in treatment response prediction, intratumor heterogeneity capturing, 13 and radiation toxicity assessment. 14 Despite this, the current radiomics workflows are designed to extract features unsuited for image segmentation due to their reliance on the predefined region of interest (ROI). Current radiomics analyses are not spatially explicit in nature. Quantitative boundary, shape, and texture features are typically generated over an ROI comprising the entire volume. 15 Additionally, current radiomics approaches assume that ROI for feature extraction is homogeneous or heterogeneous but well mixed. 16 Radiomics features extracted from the entire region cannot be used to obtain subregional quantitative image information (QII). Therefore, it is important to develop radiomics‐based methods used for segmentation to better characterize the heterogeneity within ROI.
Subregional radiomic features have been proposed to capture QII of subvolumes created by the clustering method (c‐subvolume). 17 Subregional features extracted from magnetic resonance imaging (MRI) and CT images could capture QII differences between c‐subvolumes. 18 , 19 For example, the lung tumor was classified into three c‐subvolumes (i.e., marginal subregion, fragmental subregion, and inner subregion) to detect the epidermal growth factor receptor mutation using MRI images, 19 among which inner subregion features displayed the optimum predictive performance. These results indicated the potential application of QII difference in subvolume classification. The use of the image intensity in different MRI sequences for ROI segmentation has been described. 16 However, no study has examined the feasibility of radiomic information in image segmentation.
Assuming that volumes can be segmented by grouping and merging subvolume with similar QII, this study is designed to develop a radiomics‐based framework for image segmentation (RFIS). The feature reproducibility for image segmentation is investigated using phantom images. In addition, RFIS is constructed and applied to gross target volume (GTV) delineation of lung cancers (RFIS LC ) to verify its feasibility.
2. MATERIALS AND METHODS
2.1. Study concept and design
RFIS comprised five structures: manual image labeling, svfeature extraction, svfeature selection, swvolume classification, and segmentation, as shown in Figure 1. In RFIS, features were extracted from the volume (svfeature) created by a sliding window (swvolume). Radiomics‐based feature maps could be reconstructed using svfeatures. The reproducible and non‐redundant svfeatures were selected (feature set G in Figure 1) for RFIS construction. Then, the trained RFIS was applied for the classification of unlabeled swvolumes. The swvolumes with identical classification results were grouped and merged. Post‐processing was performed to remove minimal subvolumes and create relatively large regions.
FIGURE 1.
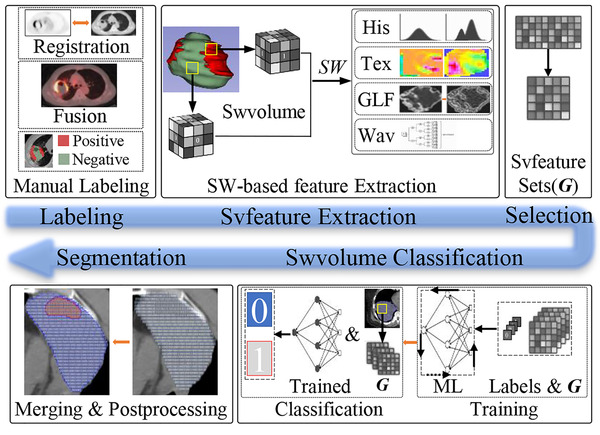
Flowchart of the RFIS procedure. His, Tex, GLF, Wav, ML, and SW denote histogram, textural, Gaussian Laplacian filtering, wavelet features, machine learning, and sliding windows, respectively. Swvolume and Svfeature mean subvolume created by the sliding window and feature extracted from swvolume, respectively
The workflow of this study consisted of two parts, as shown in Figure 2. In the first part, the reproducibility of the svfeature was assessed using phantom images. In the second part, the feasibility of RFIS was investigated using non‐small cell lung cancer (NSCLC) patient images. RFIS was constructed for the GTV segmentation from the peritumoral region (GTV with a 10 mm margin).
FIGURE 2.
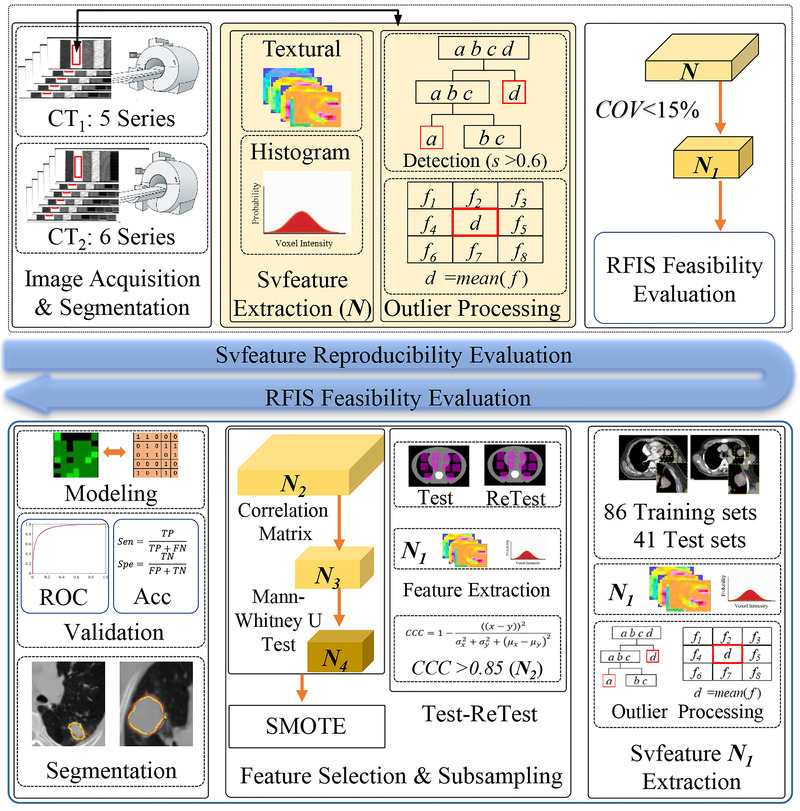
Workflow of this study. SMOTE, ROC, Acc, Sen, and Spe denote synthetic minority over‐sampling technique, receiver operating characteristic (ROC) curve, accuracy, sensitivity, and specificity, respectively. N and f mean the feature sets and svfeature value, respectively
2.2. Image datasets
Credence cartridge radiomics (CCR) phantom was designed to investigate the robustness of radiomic features. 20 The CCR phantom images acquired using different imaging protocols and scanners have been provided to The Cancer Imaging Archive (TCIA) (www.cancerimagingarchive.net) by Mackin et al. 21 , 22 CCR phantom images used in the present study were downloaded from TCIA. 21 The selected CCR images were acquired using chest protocol and were taken by six GE (General Electric Healthcare, Chicago, IL, USA) and five Philips (Philips Medical System, Netherlands) scanners. The scanning parameters contained a tube voltage of 120 kV and a slice thickness of 2.5 or 3 mm. More details about the parameters for CCR image acquisition and reconstruction can be found in Table S1.
The retrospective image database encompassed CT images from 127 NSCLC patients treated using intensity‐modulated RT or three‐dimensional (3D) conformal RT. This retrospective study was approved by the local ethics committee. All images were acquired using a Philip Brilliance Big Bore CT (Brilliance iCT 128, Philips Medical System, Netherlands) scanner and randomly divided into training sets and test sets (2:1). The scanning parameters covered a tube voltage of 120–140 kV, a tube current of 250–350 mA, and a slice thickness of 3 mm.
Anthropomorphic heterogeneous chest phantoms (AHCP, Model 002LFC, 82 CIRS, Norfolk, VA, USA) were imaged at 15‐minute intervals using Philip Brilliance Big Bore CT in our hospital to facilitate the test‐retest method. Spiral CT scans were performed using a 3 mm stacked axial slice technique with a pitch of 0.938. The resolution of those test and retest images was 512 × 512 pixels.
2.3. Image segmentation
The CCR phantom image datasets provided by Mackin et al. 22 contained a set of contours as digital imaging and communications in medicine (DICOM) RT struct files. This set provided contours of 8 × 8 × 2 cm3 for the cartridge in each scan. The cartridge of homogenous polymethyl methacrylate (acrylic) with a density of 1.1 g/cm3 was analyzed to evaluate the reproducibility of the svfeature.
For NSCLC images, radiologists delineated GTV in the lung (level, −450 HU; width, 1500 HU) and mediastinal (level, 40 HU; width, 400 HU) window settings using the Eclipse treatment planning system (Varian Medical Systems, Inc., Version 15.5, USA, TPS). The peritumoral region was defined as the GTV with a 10 mm margin, and swvolume was selected from the peritumoral region. Swvolumes with tumor voxels were labeled positive, while other swvolumes were labeled negative.
For the test‐retest images, 20 ROIs (AHCP‐ROI), including different materials, were delineated on the test images and transferred to the retest images after rigid registration using Eclipse TPS.
2.4. Svfeature extraction and outlier control
Svfeatures were extracted in MATLAB (version 9.5.0, MathWorks Inc.) using available radiomics analysis toolboxes (https://github.com/mvallieres/radiomics/) created by Vallières et al. 23 and in‐house developed software, including 53 svfeatures. These radiomics toolboxes have been used for clinical prediction tasks 24 and radiomics feature robustness evaluation, 25 , 26 bringing enormous benefits to radiomics studies. The extracted svfeatures were derived from five categories and extracted from phantom and NSCLC images. They encompassed 13 first‐order statistical (FOS) features, 9 gray‐level co‐occurrence matrix (GLCM) features, 13 gray‐level run‐length matrix (GLRLM) features, 13 gray‐level size zone matrix (GLSZM) features, and five neighboring gray‐tone difference matrix (NGTDM) features (feature set N in Figure 2). The mathematical definition of the radiomic features is listed in the Supporting Information. All the features were extracted from the original CT images, which were rescaled to 8‐bit images. All volumes were resampled to an isotropic voxel size and set to the desired resolution using cubic interpolation before feature extraction. In this study, the isotropic voxel size was scaled to 1 × 1 × 1 mm3.
The unintended inclusion of adjacent structures such as bone could affect the reliability of radiomics features. 27 Such outliers might be accidentally included in the swvolume, causing unrealistic svfeatures. The model‐based isolation forest (iForest) 28 is a simple and useful detecting method of outliers, and the number of trees t = 100 was used for isolation tree building in this study. Anomaly score (s) for each svfeature value was calculated, with s > 0.6 indicating a potential anomaly. 28 The outliers in the svfeature datasets were specified as the mean value.
2.5. Reproducibility evaluation of the Svfeatures
For the svfeatures extracted from all the CCR phantom series, the percentage coefficient of variation (%COV) was calculated to evaluate their reproducibility, as expressed in Equation (1):
| (1) |
where %COV is the percentage of COV, SD and Mean are the standard deviations. The mean of svfeatures was calculated from all the CCR image series. Svfeatures with %COV < 15 were considered reproducible ( N1 ). In addition, the average svfeature in each CCR phantom series was also calculated separately.
2.6. Feasibility evaluation of RFIS
2.6.1. Feature selection
Based on the training sets, the feature selection was conducted using a three‐step strategy to avoid model overfitting and potential bias in classifying tumor and peritumoral swvolumes. First, a test–retest method was employed to remove nonreproducible features; even measurement was carried out in images obtained from the same phantom within 15 min using the same scanner. Second, the value of Spearman's rank correlation coefficient between each pair of features was calculated as a selection tool to remove highly corrected features. Third, the Mann–Whitney U test was used to preliminarily screen the features relevant to differentiating tumor and peritumoral swvolumes.
For the test‐retest purpose, the concordance correlation coefficient (CCC) was calculated 29 for the features extracted from AHCP‐ROIs. Features with an average CCC greater than 0.85 ( N2 ) were considered reproducible and concordant. 30
The non‐redundant set of svfeatures ( N3 ) extracted from lung cancer images was selected using the correlation matrix. The column‐wise average absolute correlation was calculated for each svfeature where:
| (2) |
For each pair‐wise correlation cij exceeding 0.8, svfeatures with higher column‐wise average absolute correlation C were removed. 31 To upgrade the classification performance, Mann–Whitney U tests were conducted using SPSS (SPSS version 19, IBM) to screen out the statistical significance of svfeatures. In this univariate analysis, svfeatures ( N4 ) with p < 0.05 were considered statistically significant and were used for RFIS LC construction.
2.6.2. Subsampling
The experiment considered the imbalance between tumor and peritumoral swvolumes, inconsistent with the balanced endpoint hypothesis of most machine learning‐based classification models. The synthetic minority over‐sampling technique (SMOTE) generated synthetic minority class instances with hyperlink segments in feature space. 32 SMOTE algorithm was applied to synthesize more balanced training data with a 200% oversampling rate. It is worth noting that the synthetic new data merely appears in the training set.
2.6.3. RFIS LC construction
RFIS LC was constructed and assessed using the ten‐fold stratified cross‐validation. A support vector machine (SVM) was used to train the selected svfeatures in the training sets. The hyperparameters were optimized by Bayesian optimization using the fitcsvm function in MATLAB. Model training was performed with the Gaussian kernel function, with a kernel parameter of 1.31, a box constraint of 0.91, and a misclassification cost of 1.
The receiver operating characteristic (ROC) curve with the corresponding area under the ROC curve (AUC) was calculated to evaluate the classification performance of the swvolume, which was also assessed by the sensitivity, specificity, and accuracy, as defined in Equations (3)–(5):
| (3) |
| (4) |
| (5) |
where TP and FN are the numbers of correctly and incorrectly classified positive samples, respectively; TN and FP are the numbers of correctly and incorrectly classified negative samples, respectively. The confidence intervals of the test sets were obtained by bootstrapping the test data sets of 2000 random samples.
The swvolumes with identical predicted results were grouped and merged to determine the GTV boundary (GTV RFIS ). Window‐based mode filtering 33 , 34 (filter size: [7 7 3]) was performed to remove minimal subvolumes and create relatively large regions. The Dice similarity coefficient (DSC) represents the overlapping degree of GTV RFIS with the manually segmented GTV, as computed as follows:
| (6) |
where GTVM and GTVRFIS denote the manually delineated GTV and RFIS, respectively.
3. RESULTS
3.1. Image datasets
For the 11 CCR phantom series, 30249 swvolumes were analyzed. The mean swvolume number of the 11 CCR image series was 2749.91 ± 824.45. The swvolume numbers for each CCR phantom series can be found in Table S1.
A total of 127 NSCLC patients (85 males and 42 females, mean age: 65.27 years, range: 36–87 years) were enrolled. The 86‐image series were randomly selected for RFIS LC training, and 41 were selected for external validation. A total of 145008 swvolumes were analyzed, including 97374 (positive: negative = 23283: 74091) in the training sets and 47634 (positive: negative = 10638: 36996) in the test sets.
3.2. Reproducibility evaluation of Svfeatures
The 53 svfeatures were extracted from 30249 CCR phantom swvolumes and 49 svfeatures with %COV < 15 ( N1 ). Six svfeatures had a mean %COV < 1, seven svfeatures had a mean %COV ranging from 1 to 3, six svfeatures had a mean %COV fluctuating between 3 and 5, and 24 svfeatures had a mean %COV changing between 5 and 10, and six svfeatures had a mean %COV between 10 and 13. Four svfeatures (i.e., FOS‐Skewness, FOS‐Kurtosis, GLCM‐Correlation, GLSZM‐LZLGE) with %COV > 15 were removed in the remaining analysis steps. The variation range of the mean value of the first‐order svfeature was wider than that of the textural‐based (GLCM, GLRLM, GLSZM, and NGTDM) svfeature, as shown in Figure S1.
3.3. RFIS LC construction
3.3.1. Feature selection
For the 49 reproducible svfeatures ( N1 ), 45 features ( N2 ) had a CCC > 0.85. Four features (i.e., GLRLM‐LRLGE, GLRLM‐GLV, GLSZM‐SZLGE, and GLSZM‐LGZE) with CCC < 0.85 were considered nonreproducible and removed. The correlation matrix was used to select the non‐redundant svfeatures extracted from the NSCLC images, and 13 svfeatures ( N3 ) passed this test. The 13 non‐redundant svfeatures ( N3 ) passed the Mann–Whitney U tests (p < 0.05). Selected svfeatures for RFIS construction are shown in Table S2. These selected svfeatures from four categories and provide independent information about an image. Combinations of these categories can provide additional QII.
3.3.2. RFIS LC construction and validation
A total of 149076 samples after subsampling (positive: negative: = 72762:76314) were used for RFIS LC construction. The average accuracy of RFIS LC by the ten‐fold stratified cross‐validation was 83.48% (95% CI: 83.27%–83.70%). ROC analysis indicated that the AUC was 0.906 (95% CI: 0.904–0.908), the sensitivity was 0.848(95% CI:0.844–0.883), and the specificity was 0.821(95% CI: 0.818–0.825). The ROC curve of the training sets is shown in Figure 3a. For the independent external testing, RFIS LC achieved an accuracy of 82.29% (95% CI: 81.90%–82.60%), a sensitivity of 0.762(95% CI: 0.754–0.770), a specificity of 0.840(95% CI: 0.837–0.844), and an AUC of 0.877 (95% CI: 0.873–0.881). The ROC curve of the test sets is shown in Figure 3b.
FIGURE 3.
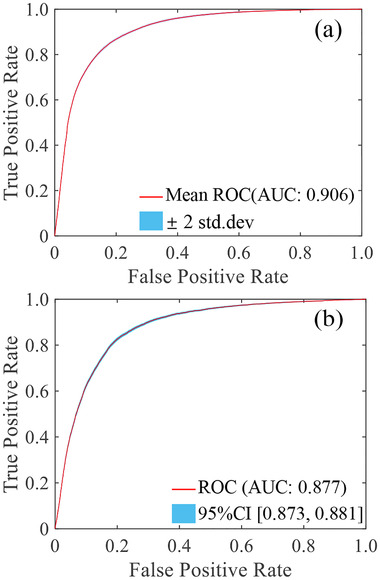
ROC curve of the training sets (a) and the test sets (b)
The CT image and corresponding svfeature map are shown in Figure 4. The mean DSC was 0.707 ± 0.093 in the training sets and 0.688 ± 0.072 in the test sets. The comparison of GTV RFIS and human expectations of six NSCLC cases is presented in Figure 5.
FIGURE 4.
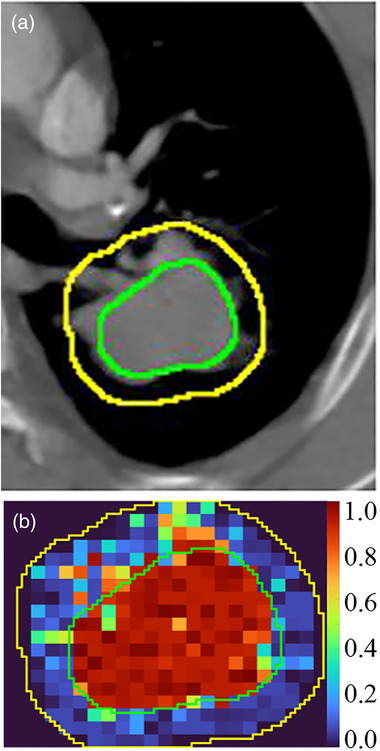
The CT images (a) and corresponding svfeature map (b). Green lines indicate gross target volume (GTV) defined by radiologists, and yellow lines denote the peritumoral region
FIGURE 5.
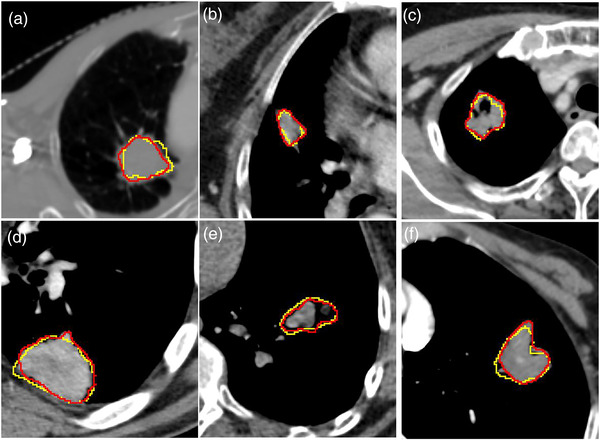
The results of GTV contours between RFIS (yellow lines) and human experts (red lines). Six cases of lung tumors with different shapes and positions
4. DISCUSSION
This study presented and verified a radiomics framework for image segmentation. Our results indicated that the svfeatures could capture the QII difference between swvolumes and be applied for image segmentation.
Outlier control is an essential factor affecting radiomics features. 35 The unintended inclusion of outliers within volumes will greatly affect GLCM and GLRLM. 27 Outlier removal (values larger or smaller than three standard deviations relative to the mean intensity value, μ ± 3σ) has been identified to increase the reproducibility of features in recent publications. 23 , 36 , 37 However, image information may be distorted while removing intensities outside the range of μ ± 3σ. In RFIS, all voxels within the image were analyzed to avoid missing QII of swvolume. Outliers were detected by iForest and specified as the mean subfeature value. Previous research 28 has identified that path lengths usually converge well before the number of trees t = 100, and s > 0.6 was a potential anomaly. Therefore, we used t = 100 and considered the cutoff value of 0.6 as an outlier.
The %COV has been used to evaluate the feature reproducibility, and features with %COV ≤ 20 were considered reproducible. 38 , 39 In our study, a stricter and more conservative %COV cutoff value (%COV < 15) was used to minimize the possibility of false positives. The textual svfeatures (GLCM, GLRLM, GLSZM, and NGTDM) showed better reproducibility and consistency than the first‐order svfeatures. The reproducibility of svfeatures after outlier control was acceptable for capturing the QII difference between swvolumes.
Radiomics features qualified as potential imaging biomarkers should be robust. The test‐retest method was used to eliminate unreproducible features. 40 In addition, radiomics analysis is plagued by feature redundancy because of the high correlation of many features. 41 The feature space was reduced into a non‐redundant subspace using correlation‐based feature elimination. The test datasets were used to avoid false‐positive results, 40 and the RFIS predictive results were found to be similar to those in the training data. The great classification accuracy for swvolume indicated the capability of svfeatures to capture QII difference between swvolumes and its applicability to swvolume classification.
The images were segmented by grouping and merging the swvolumes with similar QII in RFIS. The segmentation of minimal subvolumes was deemed undesirable. Mode filtering was applied to process 3D categorical data to remove minimal subvolumes and create relatively large regions of completely uniform character. Compared with the mean filter, the mode filter could avoid many undefined values halfway between two classes. Mode filtering causes almost no loss in boundary resolution because the filter output will not change until most of the values change. Mode filtering could improve the final texture segmentation in gray‐scale images, which has been regarded as the optimal way of discrete attribute filtering. 42 Many region sizes (3 × 3, 7 × 7, and 15 × 15) of mode filtering have been used in 2D image processing. 34 , 42 Considering the region size larger than 7 × 7 is rarely required in the mode filtering. 33 Therefore, the region size of 7 × 7 was used in the left–right and anterior–posterior axes. The superior–inferior axis resolution of CT images was coarser than that of the left–right and anterior–posterior axes. More acceptable window sizes of three voxels were used in the superior‐inferior axes.
The local entropy within the volume was calculated by moving window (9 × 9) to create the local entropy map. 18 , 19 Based on the local entropy map, ROI was divided into several c‐subvolumes using the K‐means clustering algorithm. 8 , 9 Considering that the K‐means clustering was unsupervised, the biological and/or clinical meaningfulness of the c‐subvolume was uncertain. Significantly, the biological and/or clinical meaningfulness of the c‐subvolume is usually determined by radiomics. 8 , 9 In RFIS, swvolumes were classified using supervised machine learning algorithms. Therefore, the biological and/or clinical significance of volumes segmented by RFIS was more easily confirmed. In addition, the relationship between features and tumor biology has been clarified. 43 For example, the long‐run high‐grey‐level emphasis feature extracted from CT images is positively associated with tumor hypoxia. 44 These findings are more conducive to clarify the clinical significance of volume segmented RFIS using features with clear biological connections. In addition, more high‐dimensional QII can be used in RFIS compared to unsupervised clustering algorithms, which may improve the accuracy of image segmentation.
The previous researches 45 , 46 have investigated the performance of DL‐based models in GTV segmentation for lung cancers. ResSE‐Unet trained using 148 cases achieved an optimum performance with a DSC of 0.74. 45 Similar results were obtained in another lung tumor segmentation research based on the 2D and 3D hybrid convolutional neural network (CNN) trained using 180 cases, with DSC reaching 0.72. 46 The DSC of RFIS LC was 0.707 ± 0.093 in the training sets and 0.688 ± 0.072 in the test sets. By using the DSC cut‐off values proposed by Yamamoto, 47 the similarity was interpreted as almost perfect (0.8 < DSC ≤ 1.0), substantial (0.6 < DSC ≤ 0.8), moderate (0.4 < DSC ≤ 0.6), fair (0.2 < DSC ≤ 0.4), and slight (0 < DSC ≤ 0.2). RFIS could segment substantial lung tumors, similar to the results of DL. RFIS might not compete with the current state of the art since it performed similarly to the Unet‐based methods but only segmented from the 10 mm margin around the tumor. However, the RFIS development paved the way for radiomics‐based subregional tumor segmentation. Assessing the feasibility of RFIS is the first step in the development of radiomics‐based subregional tumor segmentation, and the results of our study are sufficient to demonstrate the feasibility of RFIS. Subregional tumor segmentation is quite hard due to the unavailability of the ground truth and the greater challenges of coming up with a clean way of validating the models. RFIS might have an advantage over DL in subregional tumor segmentation since well‐formulated unsupervised methods based on radiomics can be applied.
Compared with the DL segmentation network, RFIS adopted fewer parameters and highly effective machine learning classification algorithms, 48 shortening the training time and dataset requirements. Radiomics features combined with SVM for image segmentation were beneficial to improving the generalization of the model. Radiomics features describe the distribution pattern of gray‐level pixel values (first‐order statistical features) and the spatial relationship between each pixel and its neighboring pixels (texture features). Radiomics features are less sensitive to cohort size than DL features. 49 SVM was used to perform non‐linear classification using the kernel trick that mapped to higher dimensional feature space. The hyperplane of SVM maximized the margin between the two classes in the feature space. SVM tolerated some points on the wrong side of the boundary, improving the robustness and generalization of the models. 50
The DL networks are commonly referred to as a “black box” with the internal decision processes failing to be comprehended. In the RFIS framework, mathematically defined features were designed to describe specific gray‐scale information, enabling us to clarify the gray‐scale information capable of being used to segment abnormal regions. Feature selection and shallow machine learning classification algorithms are inherently interpretable by humans. The predefined features and the machine learning‐based modeling process bring several benefits as follows. First, specialists can better understand the learning mechanism from data and the failure mechanism of the models in the new data. Second, physicians could further grasp the inner workings of the utilized tools, thus increasing their confidence in relying on the models. Moreover, svfeatures may contribute to combining radiomics and CNN for medical image segmentation. The 2D radiomics feature maps have been fed into DL models for pneumonia classification. 51 The 3D radiomics svfeature maps and medical images can be fed simultaneously into CNN for image segmentation. This approach incorporated the radiomics information into the CNN model, potentially enhancing the performance of the CNN‐based segmentation framework.
Our study has a few limitations. First, the feasibility of RFIS was investigated only in CT images. The RFIS performance should be further investigated in many other imaging modalities, such as MRI and cone‐beam CT images. Second, a few kinds of svfeatures were extracted and evaluated. More kinds of svfeatures can capture more information from swvolumes, thereby elevating the RFIS performance.
5. CONCLUSIONS
We presented and identified a radiomics framework for image segmentation. The reproducibility of svfeatures is acceptable for capturing QII within the swvolume. RFIS could be applied to swvolume classification, achieving image segmentation by grouping and merging the swvolume with similar QII.
CONFLICT OF INTEREST
The authors have no conflicts to disclose.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
This work was supported by the Shandong Provincial Natural Science Foundation under grants ZR2020LZL001 and ZR2020QH198; the National Natural Science Foundation of China under grants 81530060, 81874224, 82001902, and 81671785; the Academic promotion program of Shandong First Medical University under grants 2019LJ004 and 2020RC003; the Taishan Scholar Construction Project under grant 201909140.
Gu J, Li B, Shu H, Zhu J, Qiu Q, Bai T. Development and verification of radiomics framework for computed tomography image segmentation. Med Phys. 2022;49:6527–6537. 10.1002/mp.15904
Contributor Information
Baosheng Li, Email: bsli@sdfmu.edu.cn.
Huazhong Shu, Email: shu.list@seu.edu.cn.
DATA AVAILABILITY STATEMENT
Data is available from the corresponding author (B.S.L.) upon request.
REFERENCES
- 1. Peeken J, Nüsslin F, Combs S. “Radio‐oncomics”: the potential of radiomics in radiation oncology. Strahlenther Onkol. 2017;193(10):767‐779. 10.1007/s00066-017-1175-0 [DOI] [PubMed] [Google Scholar]
- 2. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5(1):1‐9. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brouwer CL, Steenbakkers RJ, van den Heuvel E, et al. 3D variation in delineation of head and neck organs at risk. Radiat Oncol. 2012;7(1):1‐10. 10.1186/1748-717X-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walker GV, Awan M, Tao R, et al. Prospective randomized double‐blind study of atlas‐based organ‐at‐risk autosegmentation‐assisted radiation planning in head and neck cancer. Radiother Oncol. 2014;112(3):321‐325. 10.1016/j.radonc.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kosmin M, Ledsam J, Romera‐Paredes B, et al. Rapid advances in auto‐segmentation of organs at risk and target volumes in head and neck cancer. Radiother Oncol. 2019;135:130‐140. 10.1016/j.radonc.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 6. Chen X, Wang X, Zhang K, et al. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal. 2022;79:102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U‐Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W, eds. Medical Image Computing and Computer‐Assisted Intervention‐MICCAI 2016. Springer; 2016:424‐432. [Google Scholar]
- 8. Syed AK, Whisenant JG, Barnes SL, Sorace AG, Yankeelov TE. Multiparametric analysis of longitudinal quantitative MRI data to identify distinct tumor habitats in preclinical models of breast cancer. Cancers. 2020;12(6):1682. 10.3390/cancers12061682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shang S, Sun J, Yue Z, Wang Y, Jiang X. Multi‐parametric MRI based radiomics with tumor subregion partitioning for differentiating benign and malignant soft‐tissue tumors. Biomed Signal Process Control. 2021;67(8):102522. 10.1016/j.bspc.2021.102522 [DOI] [Google Scholar]
- 10. Lambin P, Leijenaar RT, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749‐762. 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 11. Chen J, Zeng H, Zhang C, et al. Lung cancer diagnosis using deep attention‐based multiple instance learning and radiomics. Med Phys. 2022;49(5):3134‐3143. 10.1002/mp.15539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avanzo M, Gagliardi V, Stancanello J, et al. Combining computed tomography and biologically effective dose in radiomics and deep learning improves prediction of tumor response to robotic lung stereotactic body radiation therapy. Med Phys. 2021;48(10):6257‐6269. 10.1002/mp.15178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanduleanu S, Jochems A, Upadhaya T, et al. Non‐invasive imaging prediction of tumor hypoxia: a novel developed and externally validated CT and FDG‐PET‐based radiomic signatures. Radiother Oncol. 2020;153:97‐105. 10.1016/j.radonc.2020.10.016 [DOI] [PubMed] [Google Scholar]
- 14. Cunliffe A, Armato SG III, Castillo R, Pham N, Guerrero T, Al‐Hallaq HA. Lung texture in serial thoracic computed tomography scans: correlation of radiomics‐based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol Biol Phys. 2015;91(5):1048‐1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walker‐Samuel S, Orton M, Boult JK, Robinson SP. Improving apparent diffusion coefficient estimates and elucidating tumor heterogeneity using Bayesian adaptive smoothing. Magn Reson Med . 2011;65(2):438‐447. 10.1002/mrm.22572 [DOI] [PubMed] [Google Scholar]
- 16. Gatenby R, Grove O, Gillies R. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8‐15. 10.1148/radiol.13122697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang T, Jiang W, Chang S, et al. Intratumoral analysis of digital breast tomosynthesis for predicting the Ki‐67 level in breast cancer: a multi‐center radiomics study. Med Phys. 2021;49(1):219‐230. 10.1002/mp.15392 [DOI] [PubMed] [Google Scholar]
- 18. Xie C, Yang P, Zhang X, et al. Sub‐region based radiomics analysis for survival prediction in oesophageal tumours treated by definitive concurrent chemoradiotherapy. EBioMedicine. 2019;44:289‐297. 10.1016/j.ebiom.2019.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan Y, Dong Y, Yang H, et al. Subregional radiomics analysis for the detection of the EGFR mutation on thoracic spinal metastases from lung cancer. Phys Med Biol. 2021;66(21). 10.1088/1361-6560/ac2ea7 [DOI] [PubMed] [Google Scholar]
- 20. Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50(11):757. 10.1097/RLI.0000000000000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clark K, Vendt B, Smith K, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045‐57. 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. FX Mackin Dennis, Lifei Zhang, David Fried, et al. Data from credence cartridge radiomics phantom CT scans. The Cancer Imaging Archive. 2007. 10.7937/K9/TCIA.2017.zuzrml5b [DOI] [Google Scholar]
- 23. Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG‐PET and MRI texture features for the prediction of lung metastases in soft‐tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471. 10.1088/0031-9155/60/14/5471 [DOI] [PubMed] [Google Scholar]
- 24. Zhou H, Vallières M, Bai HX, et al. MRI features predict survival and molecular markers in diffuse lower‐grade gliomas. Neuro Oncol. 2017;19(6):862‐870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lv W, Yuan Q, Wang Q, et al. Robustness versus disease differentiation when varying parameter settings in radiomics features: application to nasopharyngeal PET/CT. Eur Radiol. 2018;28(8):3245‐3254. 10.1007/s00330-018-5343-0 [DOI] [PubMed] [Google Scholar]
- 26. Lu L, Lv W, Jiang J, et al. Robustness of radiomic features in [11C] choline and [18F] FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol. 2016;18(6):935‐945. 10.1007/s11307-016-0973-6 [DOI] [PubMed] [Google Scholar]
- 27. Park BW, Kim JK, Heo C, Park KJ. Reliability of CT radiomic features reflecting tumour heterogeneity according to image quality and image processing parameters. Sci Rep . 2020;10(1):1‐13. 10.1038/s41598-020-60868-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu FT, Ting KM, Zhou ZH. Isolation‐based anomaly detection. ACM Trans Knowl Discovery Data. 2012;6(1):1‐39. 10.1145/2133360.2133363 [DOI] [Google Scholar]
- 29. Balagurunathan Y, Kumar V, Gu Y, et al. Test–retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27(6):805‐823. 10.1007/s10278-014-9716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaudet C, Weyts K, Lechervy A, Batalla A, Bardet S, Corroyer‐Dulmont A. The impact of artificial intelligence CNN based denoising on FDG PET radiomics. Front Oncol. 2021;11:692973. 10.3389/fonc.2021.692973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu W, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. 2016;6:71. 10.3389/fonc.2016.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over‐sampling technique. J Artif Intell Res. 2002;16(1):321‐357. 10.1613/jair.953 [DOI] [Google Scholar]
- 33. Hall M. Smooth operator: smoothing seismic interpretations and attributes. Lead Edge. 2007;26(1):16‐20. 10.1190/1.2431821 [DOI] [Google Scholar]
- 34. Coleman GB, Andrews HC. Image segmentation by clustering. Proc IEEE. 1979;67(5):773‐785. 10.1109/PROC.1979.11327 [DOI] [Google Scholar]
- 35. Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and generalizability in radiomics modeling: possible strategies in radiologic and statistical perspectives. Korean J Radiol. 2019;20(7):1124‐1137. 10.3348/kjr.2018.0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22(1):81‐91. 10.1016/j.mri.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 37. Traverso A, Kazmierski M, Welch ML, et al. Sensitivity of radiomic features to inter‐observer variability and image pre‐processing in apparent diffusion coefficient (ADC) maps of cervix cancer patients. Radiother Oncol. 2020;143:88‐94. 10.1016/j.radonc.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 38. Shafiq‐Ul‐Hassan M, Zhang G, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44(3):1050‐1062. 10.1002/mp.12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shiri I, Rahmim A, Ghaffarian P, Geramifar P, Abdollahi H, Bitarafan‐Rajabi A. The impact of image reconstruction settings on 18F‐FDG PET radiomic features: multi‐scanner phantom and patient studies. Eur Radiol. 2017;27(11):4498‐4509. 10.1007/s00330-017-4859-z [DOI] [PubMed] [Google Scholar]
- 40. Yip S, Aerts H. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150‐66. 10.1088/0031-9155/61/13/r150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orlhac F, Soussan M, Maisonobe J, Garcia C, Vanderlinden B, Buvat I. Tumor texture analysis in 18F‐FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med. 2014;55(3):414‐22. 10.2967/jnumed.113.129858 [DOI] [PubMed] [Google Scholar]
- 42. Martins DC, Cesar RM, Barrera J. Automatic window design for gray‐scale image processing based on entropy minimization. In: Sanfeliu A, Cortes ML, eds. Progress in Pattern Recognition, Image Analysis and Applications. Springer; 2005:813‐824. [Google Scholar]
- 43. Zhang T, Xu Z, Liu G, et al. Simultaneous identification of EGFR, KRAS, ERBB2, and TP53 mutations in patients with non‐small cell lung cancer by machine learning‐derived three‐dimensional radiomics. Cancers. 2021;13(8):1814. 10.3390/cancers13081814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Crispin‐Ortuzar M, Apte A, Grkovski M, et al. Predicting hypoxia status using a combination of contrast‐enhanced computed tomography and [F]‐Fluorodeoxyglucose positron emission tomography radiomics features. Radiother Oncol. 2018;127(1):36‐42. 10.1016/j.radonc.2017.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu X, Jin F, Luo H, Lei Q, Wu Y. Gross tumor volume segmentation for stage III NSCLC radiotherapy using 3D ResSE‐Unet. Technol Cancer Res Treat. 2022;21(5):15330338221090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gan W, Wang H, Gu H, et al. Automatic segmentation of lung tumors on CT images based on a 2D & 3D hybrid convolutional neural network. Br J Radiol. 2021;94(1126):20210038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamamoto T, Kabus S, Von Berg J, et al. Reproducibility of four‐dimensional computed tomography‐based lung ventilation imaging. Acad Radiol. 2012;19(12):1554‐1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Avanzo M, Wei L, Stancanello J, et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47(5):e185‐e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parmar C, Barry JD, Hosny A, Quackenbush J, Aerts HJ. Data analysis strategies in medical imaging. Clin Cancer Res. 2018;24(15):3492‐3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen S, Zhou S, Yin FF, Marks LB, Das SK. Investigation of the support vector machine algorithm to predict lung radiation‐induced pneumonitis. Med Phys. 2007;34(10):3808‐3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu Z, Yang Z, Lafata K, Yin F, Wang C. A radiomics‐boosted deep‐learning model for COVID‐19 and non‐COVID‐19 pneumonia classification using chest x‐ray images. Med Phys. 2022;49(5):3213‐3222. 10.1002/mp.15582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Data is available from the corresponding author (B.S.L.) upon request.


