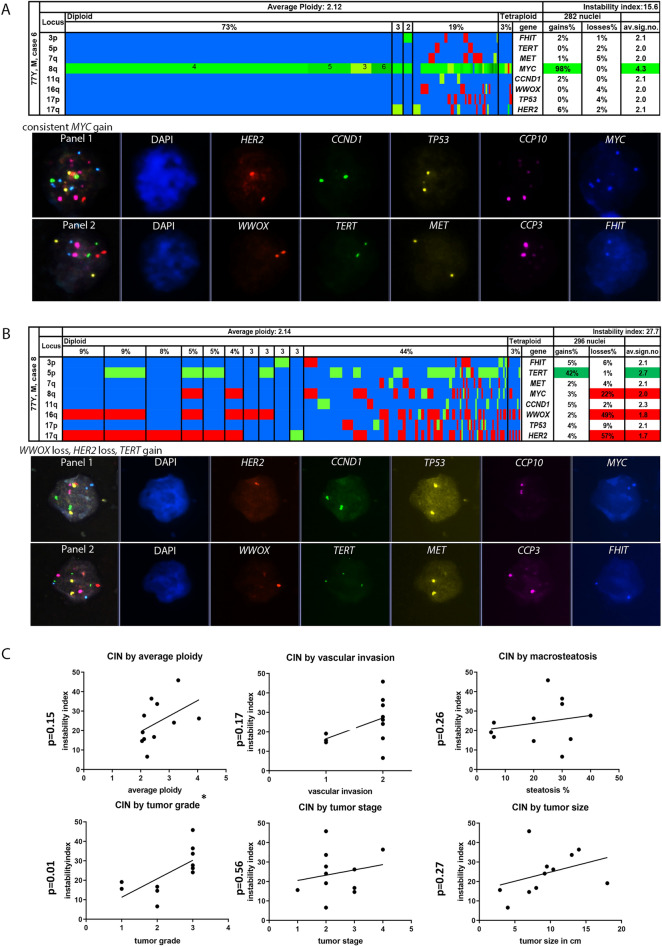
Figure 2.
(A–C) Color display of miFISH analysis with eight gene-specific probes supplemented with a gallery view of signal patterns for representative nuclei in NASH-hepatocellular carcinoma cases. Copy number counts for each nucleus coded as follows: green, gains; red, losses; blue, unchanged. Detail from left to right: the “locus” column shows the chromosome arm the probes reside on. Cells with the same gain and loss patterns are grouped according to frequency and ploidy (vertical lines). The average ploidy (center, centromere signal/number of nuclei) and the instability index (top right, number of signal patterns/100 nuclei.) are calculated from all counted nuclei (296 and 282 respectively). (A) HCC case showing consistent MYC gain in 98% in diploid and tetraploid tumor cells (color figure) and relatively low instability index of 15.6 (number of signal patterns/100 nuclei). (B) HCC case with several tumor subclones, present in diploid and tetraploid cells, high instability index (27.7), due to different signal patterns comprising HER2, WWOX loss and TERT gain. The loss of oxidoreductase WWOX and HER2 gains are potentially associated with NASH-induced hepatocellular carcinoma. DUET/SOLO acquisition and analysis software version 3.7.2.5 https://bioview.com/ (C) Correlations of histopathological findings and chromosomal instability index with respective p-values (graphPad Prism software version 9.4.1 https://www.graphpad.com/scientific-software/prism/). CIN Chromosomal instability.