Fig. 2.
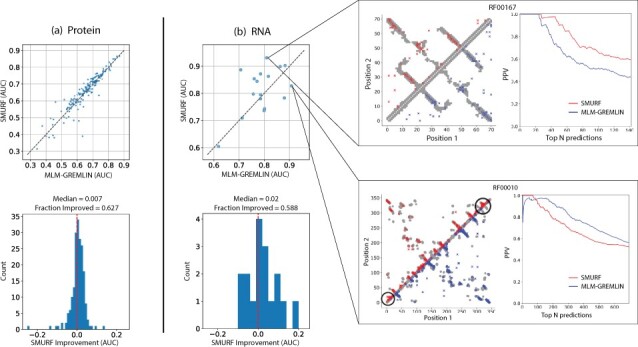
SMURF often outperforms MLM-GREMLIN on (a) protein and (b) non-coding RNA. (Top) Scatter plots of the AUC of the top L predicted contacts for SMURF versus MLM-GREMLIN. (Bottom) Histograms of the difference in AUC between SMURF and MLM-GREMLIN. (Right) Comparison of contact predictions and the positive predictive value (PPV) for different numbers of top N predicted contacts, with N ranging from 0 to 2L, for SMURF (red, above the diagonal) and MLM-GREMLIN (blue, below the diagonal) for Rfam family RF00010 (Ribonuclease P.) and RF00167 (Purine riboswitch). Gray dots represent PDB-derived contacts, circles represent a true positive prediction, and x represents a false positive prediction. For contact predictions for RFAM00010, the black circles highlight a concentration of false positive predictions (A color version of this figure appears in the online version of this article)
