Fig. 1.
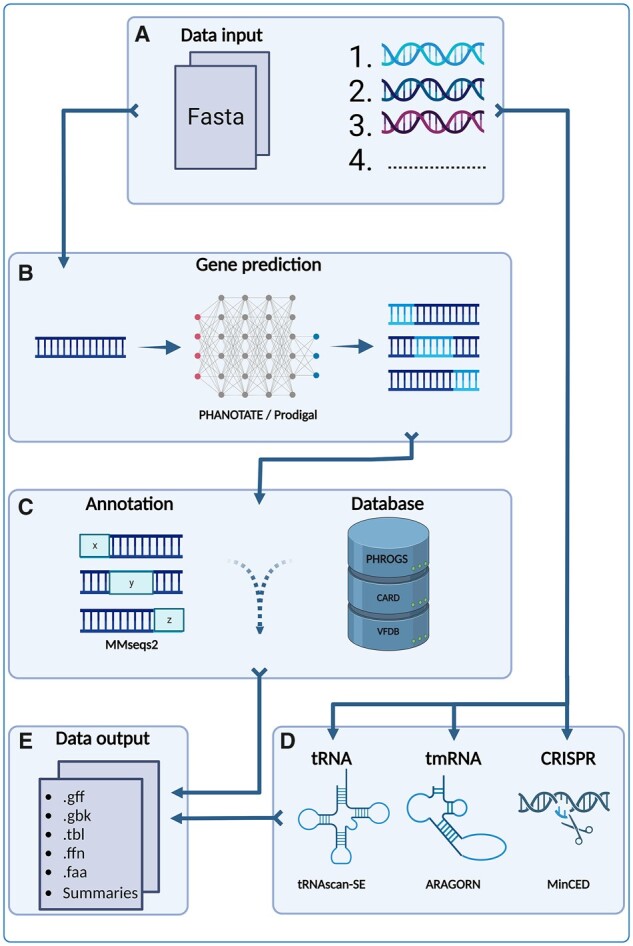
Pharokka workflow. (A) An input phage complete assembly or input phage contigs are loaded. (B) CDS are predicted with PHANOTATE (default) or Prodigal. (C) CDS are functionally annotated by matching them to the PHROGs database with mmseqs2. The CDS are then matched to the CARD and the VFDB. (D) tRNAs, tmRNAs and CRISPRs are detected using tRNAscan-SE, Aragorn and MinCED. (E) All annotations are amalgamated into standards compliant output formats
