Figure 4.
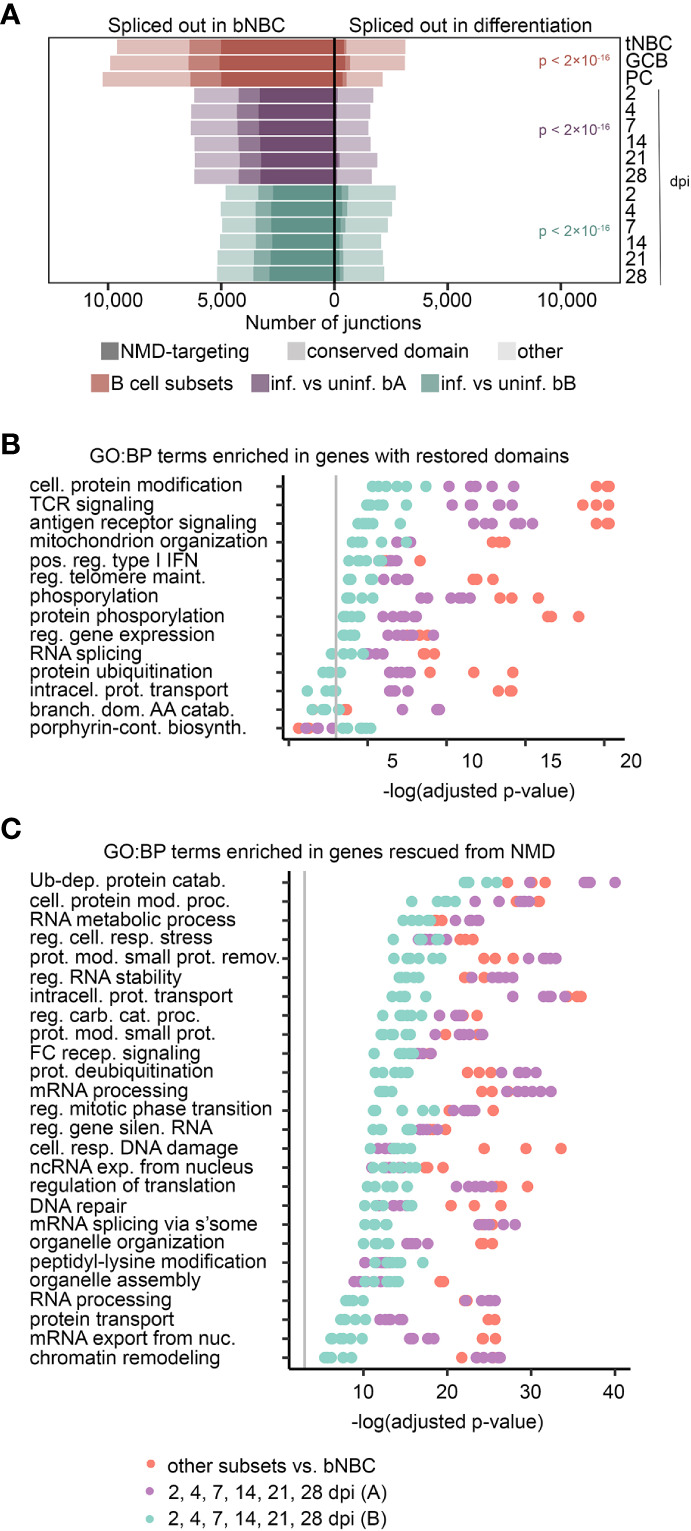
Exon inclusion restores domains and evades NMD in important pathways. (A) Exons with altered usage (rMATS FDR < 0.05) in lymphoid (tonsil) NBCs (tNBC), GC B cells (GCB) and PCs compared to blood NBCs (red) or EBV infection of blood B cells (purple = infection timecourse A, green = infection timecourse B). Exons that encode conserved protein domains are lightly shaded, exons that induce NMD targeting when skipped are heavily shaded. P-values for the proportion of damaging exon skipping events (NMD-targeting or domain-skipping) in blood NBCs compared to differentiating or EBV-infected cells were calculated using a pairwise comparison of proportions test with Bonferroni correction. (B) GO Biological Process term enrichment in genes with conserved protein domains restored by exon inclusion in differentiating or EBV-infected cells. The top 5 terms with lowest adjusted p-value for each experiment/timepoint are shown. (C) GO Biological Process term enrichment in genes with NMD targeting reduced by exon inclusion in differentiated or EBV-infected cells. The top 10 terms with lowest adjusted p-value for each experiment/timepoint are shown. For panels (B, C), redundant terms are not displayed, complete enrichment information is available in Supplemental Table 7 . Vertical grey line indicates adjusted p-value = 0.05. Adjusted p-values calculated by Enrichr using Fisher’s exact test with Benjamini-Hochberg correction. For all panels B cell subset comparisons include 3 subsets (tonsil NBCs, GC B cells, and PCs) compared to blood NBCs. Each B cell subset includes 3 replicates. Blood infection timecourse A (purple) includes 6 timepoint comparisons with 3 replicates each, Blood infection timecourse B (green) includes 6 timepoint comparisons with 1 replicate each.
